suppressor of cytokine signaling 4
ZFIN 
Other cell groups


all cells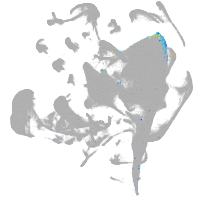 |
blastomeres |
PGCs |
endoderm |
axial mesoderm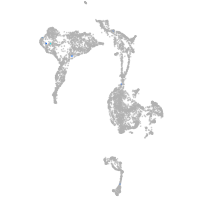 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
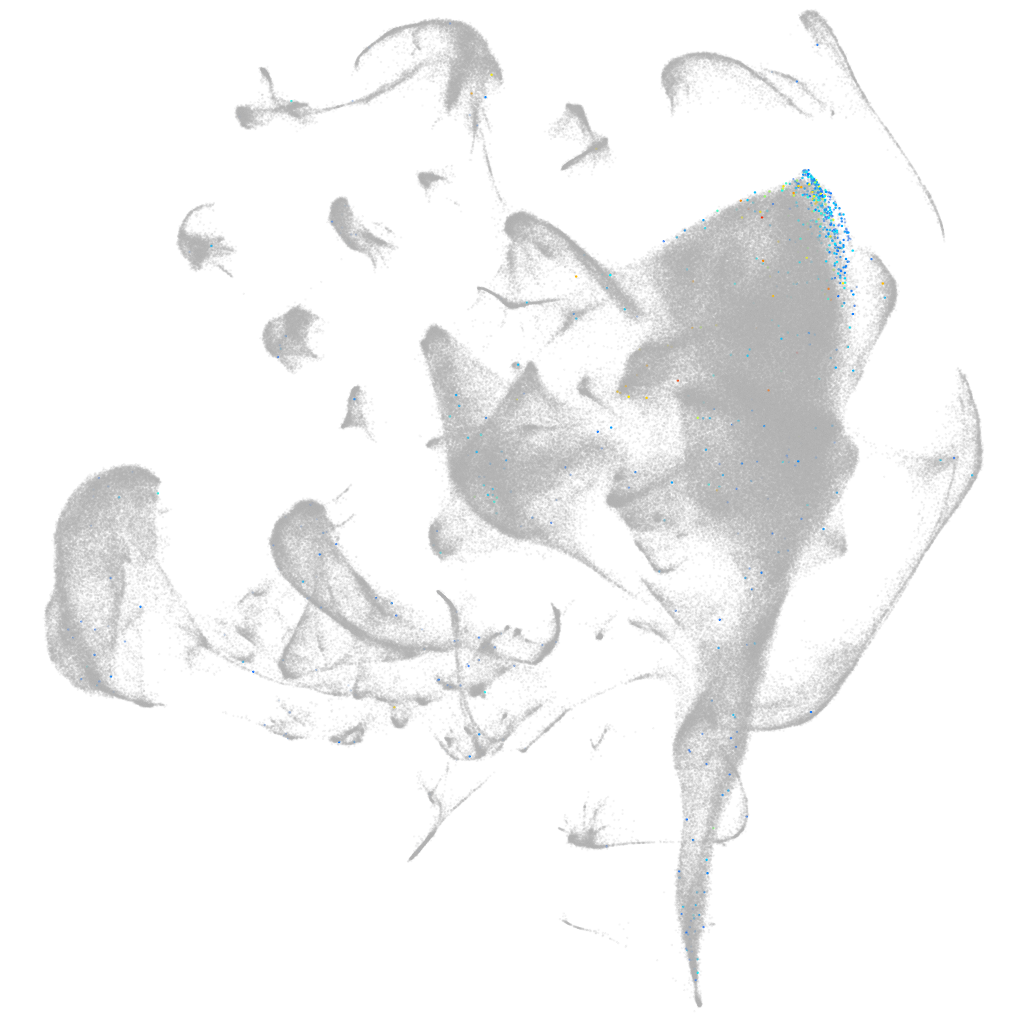
neural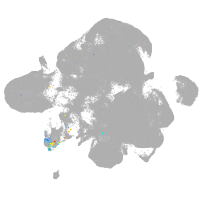 |
spinal cord / glia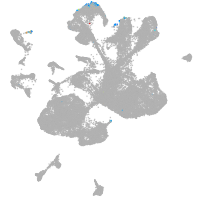 |
eye |
taste / olfactory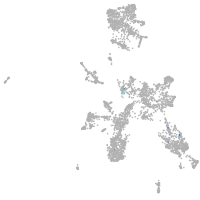 |
otic / lateral line |
epidermis |
periderm |
fin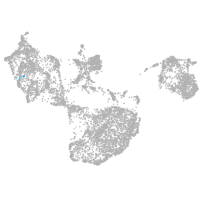 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mmp25b | 0.131 | gpm6aa | -0.015 |
| si:dkey-43k4.5 | 0.129 | celf2 | -0.012 |
| lgi2b | 0.128 | pvalb1 | -0.012 |
| prph | 0.124 | pvalb2 | -0.012 |
| ntrk1 | 0.114 | CU467822.1 | -0.011 |
| prkg2 | 0.108 | hbbe1.3 | -0.011 |
| emb | 0.104 | actc1b | -0.010 |
| tubb2 | 0.103 | hbae1.1 | -0.010 |
| sst6 | 0.099 | hbae3 | -0.010 |
| ank2a | 0.096 | mdka | -0.010 |
| anxa13l | 0.095 | mylz3 | -0.010 |
| nefma | 0.094 | slc6a1a | -0.010 |
| stmn3 | 0.092 | slc6a1b | -0.010 |
| tagln3b | 0.089 | syt5b | -0.010 |
| tspan2b | 0.086 | CU634008.1 | -0.010 |
| six2b | 0.084 | btg2 | -0.009 |
| CU693471.1 | 0.081 | eef1da | -0.009 |
| pcbp4 | 0.081 | foxp4 | -0.009 |
| prkg1a | 0.080 | hbae1.3 | -0.009 |
| ngfrb | 0.079 | hbbe1.1 | -0.009 |
| phox2a | 0.078 | hbbe2 | -0.009 |
| tlx2 | 0.078 | krt4 | -0.009 |
| nefmb | 0.077 | nr2f1a | -0.009 |
| scn1laa | 0.077 | rs1a | -0.009 |
| si:dkey-10f21.4 | 0.077 | si:dkey-16p21.8 | -0.009 |
| trarg1b | 0.077 | ahnak | -0.008 |
| ngfra | 0.076 | aldob | -0.008 |
| ppp1r1c | 0.076 | cahz | -0.008 |
| vim | 0.076 | cx43.4 | -0.008 |
| CABZ01118738.1 | 0.075 | cyt1 | -0.008 |
| capn1b | 0.075 | fstl1b | -0.008 |
| neflb | 0.075 | icn | -0.008 |
| sgk494a | 0.075 | icn2 | -0.008 |
| amigo3 | 0.074 | lhx1a | -0.008 |
| eya4 | 0.074 | mylpfa | -0.008 |