NDUFA4 mitochondrial complex associated like 2a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 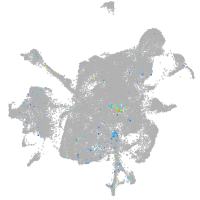 |
hematopoietic / vasculature 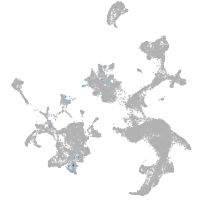 |
pronephros 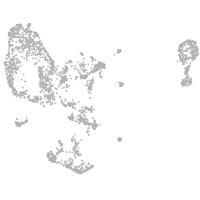 |
neural |
spinal cord / glia |
eye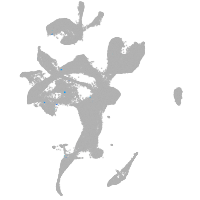 |
taste / olfactory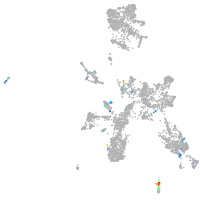 |
otic / lateral line |
epidermis |
periderm |
fin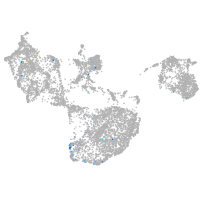 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| drd1a | 0.575 | edf1 | -0.042 |
| lama2 | 0.474 | qkia | -0.040 |
| CR759810.1 | 0.465 | sdcbp2 | -0.040 |
| XLOC-001430 | 0.453 | cirbpa | -0.039 |
| si:ch73-103l1.2 | 0.447 | mt-co2 | -0.038 |
| wfs1a | 0.416 | ssr2 | -0.038 |
| cfi | 0.382 | lsm12b | -0.037 |
| calcrla | 0.334 | pfn2l | -0.036 |
| dhrs7cb | 0.331 | pabpc1a | -0.036 |
| cfap300 | 0.306 | fth1a | -0.035 |
| hmha1a | 0.299 | tmed10 | -0.035 |
| kcne4 | 0.292 | mt-cyb | -0.034 |
| si:ch73-261i21.2 | 0.280 | nop53 | -0.033 |
| cables2b | 0.268 | atp1b1a | -0.033 |
| CR855375.1 | 0.255 | dnaja2a | -0.031 |
| LOC110440078 | 0.251 | pbx4 | -0.030 |
| BX510657.1 | 0.249 | gspt1l | -0.030 |
| tm4sf18 | 0.247 | kdelr2a | -0.030 |
| pcdh20.1 | 0.246 | ssr1 | -0.029 |
| pear1 | 0.246 | sap18 | -0.029 |
| exoc3l1 | 0.246 | srrm2 | -0.028 |
| lmod3 | 0.246 | srsf11 | -0.028 |
| LOC100330348 | 0.244 | tia1l | -0.028 |
| si:ch73-160i9.2 | 0.237 | csnk2b | -0.028 |
| CR383669.7 | 0.237 | nop58 | -0.028 |
| gcgb | 0.233 | zmat2 | -0.028 |
| chrnb3b | 0.231 | syncrip | -0.028 |
| grnas | 0.228 | hdlbpa | -0.028 |
| XLOC-025875 | 0.226 | fbl | -0.028 |
| CR735102.1 | 0.225 | pcbp2 | -0.028 |
| si:ch211-76l23.4 | 0.220 | sarnp | -0.027 |
| CR394572.1 | 0.219 | hnrnpl2 | -0.027 |
| LOC103911269 | 0.218 | ctnnb1 | -0.027 |
| pcdhb | 0.217 | cycsb | -0.027 |
| BX571724.1 | 0.215 | rsl1d1 | -0.027 |