Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line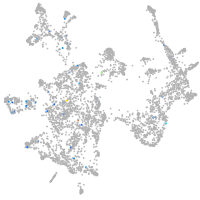 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.129 | krtt1c19e | -0.012 |
| cfap126 | 0.128 | actc1b | -0.011 |
| si:ch211-155m12.5 | 0.125 | pvalb2 | -0.011 |
| si:ch211-163l21.7 | 0.124 | si:ch211-105c13.3 | -0.011 |
| spaca9 | 0.122 | aep1 | -0.010 |
| ttc25 | 0.122 | cyt1 | -0.010 |
| ccdc151 | 0.119 | hrc | -0.010 |
| foxj1a | 0.118 | icn2 | -0.010 |
| spag6 | 0.117 | krt4 | -0.010 |
| armc3 | 0.112 | pvalb1 | -0.010 |
| tbata | 0.110 | si:ch211-195b11.3 | -0.010 |
| ccdc169 | 0.109 | wu:fa03e10 | -0.010 |
| rsph9 | 0.108 | wu:fb18f06 | -0.010 |
| nme5 | 0.103 | zgc:193505 | -0.010 |
| smkr1 | 0.103 | agr1 | -0.009 |
| morn3 | 0.102 | anxa1c | -0.009 |
| enkur | 0.101 | caspb | -0.009 |
| ccdc173 | 0.100 | col1a1b | -0.009 |
| si:dkey-27p23.3 | 0.100 | col1a2 | -0.009 |
| spag8 | 0.100 | cyt1l | -0.009 |
| dnaaf1 | 0.099 | hbae1.1 | -0.009 |
| ak7a | 0.098 | hbbe1.3 | -0.009 |
| cfap52 | 0.098 | krt17 | -0.009 |
| pih1d3 | 0.098 | krt5 | -0.009 |
| ccdc114 | 0.097 | lye | -0.009 |
| rsph4a | 0.097 | mgst1.2 | -0.009 |
| dydc2 | 0.096 | pnp5a | -0.009 |
| efcab2 | 0.095 | ppl | -0.009 |
| ribc2 | 0.095 | pycard | -0.009 |
| CFAP77 | 0.094 | s100v2 | -0.009 |
| dnali1 | 0.094 | scel | -0.009 |
| si:dkey-114c15.5 | 0.094 | scrt2 | -0.009 |
| ccdc65 | 0.093 | si:ch211-125o16.4 | -0.009 |
| si:ch211-163l21.10 | 0.093 | si:ch211-157c3.4 | -0.009 |
| tekt1 | 0.093 | si:ch73-52f15.5 | -0.009 |