myelin regulatory factor
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 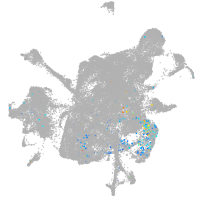 |
hematopoietic / vasculature 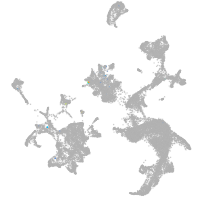 |
pronephros 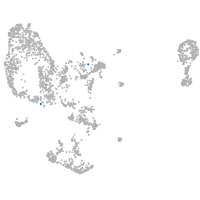 |
neural |
spinal cord / glia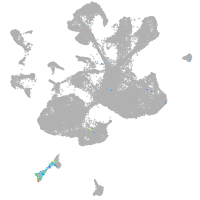 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.252 | cycsb | -0.044 |
| krt94 | 0.229 | uqcrh | -0.043 |
| tmem88b | 0.198 | npm1a | -0.036 |
| lurap1 | 0.190 | COX5B | -0.033 |
| tmem98 | 0.189 | nop56 | -0.032 |
| XLOC-013634 | 0.178 | c1qbp | -0.031 |
| cyp8b3 | 0.178 | nhp2 | -0.031 |
| ftr04 | 0.174 | hsp90aa1.1 | -0.031 |
| cavin1b | 0.173 | fbl | -0.031 |
| CU929037.3 | 0.171 | nop58 | -0.031 |
| sostdc1a | 0.166 | dkc1 | -0.030 |
| jam2b | 0.157 | ybx1 | -0.030 |
| si:dkeyp-97e7.9 | 0.156 | cox7b | -0.029 |
| cavin2b | 0.156 | rrp1 | -0.029 |
| gdf7 | 0.152 | rrs1 | -0.029 |
| cav1 | 0.147 | znf593 | -0.029 |
| ftr82 | 0.146 | lyar | -0.029 |
| crygm2d4 | 0.142 | ncl | -0.029 |
| FRMD1 | 0.140 | snu13b | -0.029 |
| c1qtnf2 | 0.136 | cox7a2l | -0.028 |
| CABZ01002809.1 | 0.136 | pabpc4 | -0.028 |
| pcolcea | 0.134 | mrpl54 | -0.028 |
| agrp | 0.132 | atp5md | -0.028 |
| BX005154.1 | 0.130 | hspe1 | -0.028 |
| rhag | 0.128 | srprb | -0.027 |
| ccl44 | 0.123 | nop10 | -0.027 |
| hand2 | 0.123 | ttn.1 | -0.027 |
| tbx5a | 0.123 | bysl | -0.027 |
| slit3 | 0.123 | cox6b2 | -0.027 |
| ikzf2 | 0.121 | ndufab1b | -0.027 |
| tmem88a | 0.120 | tuba8l2 | -0.027 |
| tmem108 | 0.120 | mrto4 | -0.027 |
| trim35-25 | 0.120 | si:dkey-239h2.3 | -0.027 |
| sema3d | 0.118 | nop16 | -0.027 |
| si:dkey-102c8.3 | 0.118 | BX927258.1 | -0.027 |




