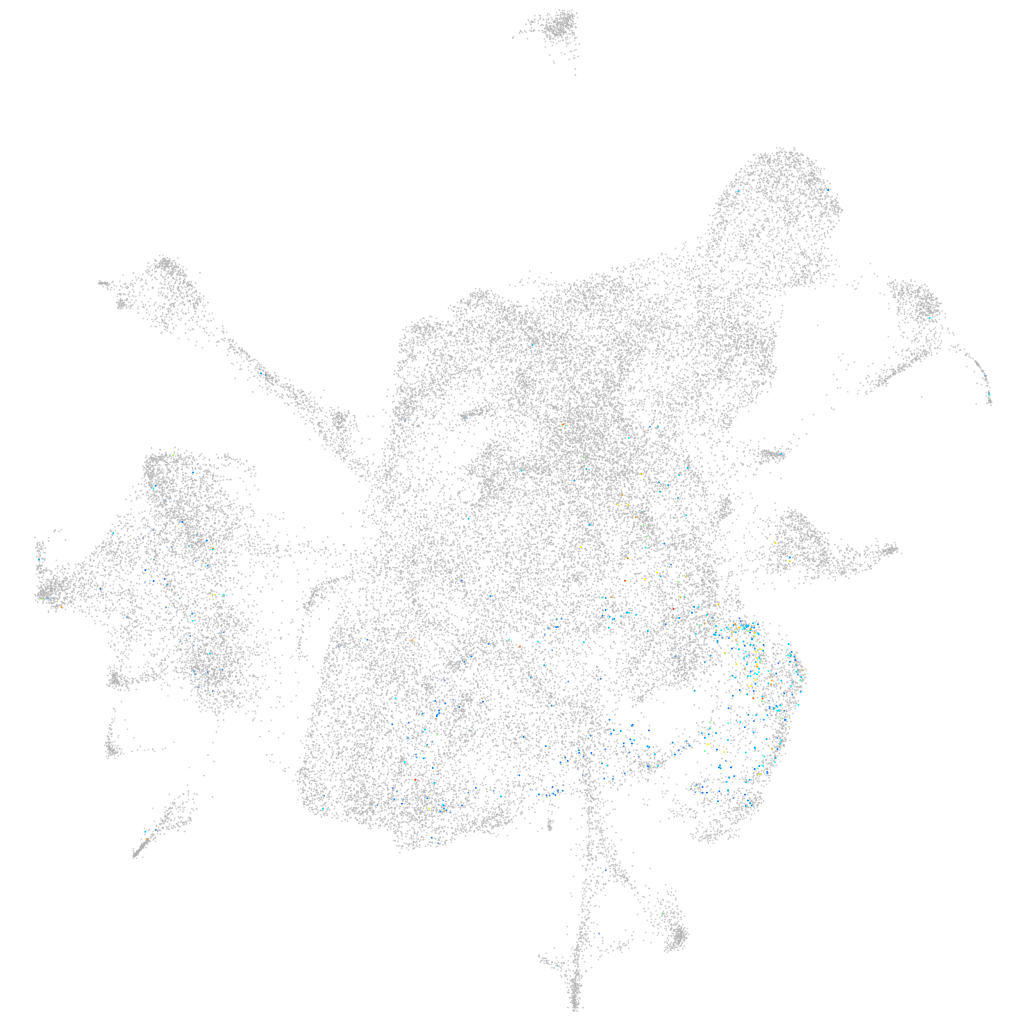
myelin regulatory factor
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 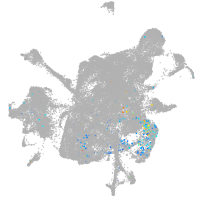 |
hematopoietic / vasculature 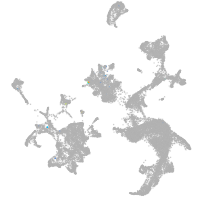 |
pronephros 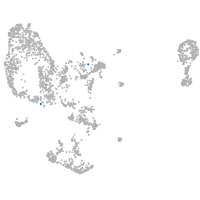 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.261 | fibina | -0.060 |
| jam2b | 0.199 | tgfbi | -0.049 |
| tmem98 | 0.183 | hapln1a | -0.048 |
| tmem88b | 0.172 | cnmd | -0.047 |
| cavin2b | 0.166 | col9a1a | -0.047 |
| agrp | 0.160 | cthrc1a | -0.045 |
| aqp1a.1 | 0.149 | matn4 | -0.045 |
| wt1a | 0.149 | tnc | -0.044 |
| fat2 | 0.148 | col9a2 | -0.044 |
| meis3 | 0.148 | rarga | -0.043 |
| gcga | 0.139 | cdh11 | -0.042 |
| rbpms2b | 0.137 | col9a3 | -0.042 |
| aldh1a2 | 0.134 | col2a1a | -0.042 |
| arl4aa | 0.131 | col2a1b | -0.041 |
| rhag | 0.129 | tnn | -0.041 |
| LOC110438194 | 0.127 | CU929237.1 | -0.039 |
| CABZ01045212.1 | 0.127 | col11a1b | -0.039 |
| cxcl8b.1 | 0.126 | fgfbp2b | -0.039 |
| ppp1r14aa | 0.121 | ecrg4a | -0.038 |
| cxcl8b.3 | 0.120 | sox9a | -0.037 |
| cd151 | 0.120 | cd248b | -0.036 |
| gata5 | 0.119 | dlx5a | -0.035 |
| ftr82 | 0.119 | cspg4 | -0.034 |
| cavin1b | 0.116 | eya1 | -0.033 |
| cav1 | 0.113 | col11a2 | -0.032 |
| lurap1 | 0.113 | six2a | -0.032 |
| zgc:110182 | 0.109 | zgc:66479 | -0.031 |
| tmem108 | 0.108 | fxyd1 | -0.031 |
| serpine3 | 0.106 | spon2b | -0.031 |
| rdh10a | 0.106 | ebf1a | -0.030 |
| alkal2b | 0.105 | six1a | -0.030 |
| tmem88a | 0.103 | phlda3 | -0.030 |
| pcdh10b | 0.100 | twist2 | -0.029 |
| gata6 | 0.099 | VIT | -0.029 |
| hspb1 | 0.098 | dlx3b | -0.029 |