major histocompatibility complex class I ZBA
ZFIN 
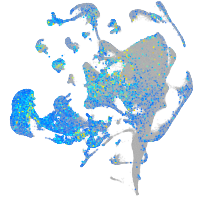
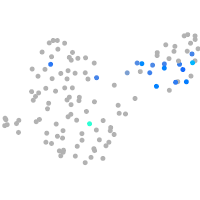
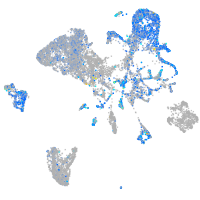
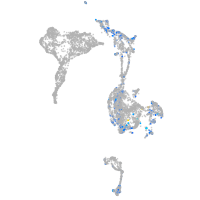
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-137i24.10 | 0.405 | bhmt | -0.315 |
| krt4 | 0.399 | aqp12 | -0.300 |
| tmem176l.2 | 0.389 | apoc1 | -0.267 |
| tmem45b | 0.387 | agxtb | -0.246 |
| cap1 | 0.385 | kng1 | -0.226 |
| cd63 | 0.385 | mat1a | -0.223 |
| serpinb1 | 0.383 | grhprb | -0.222 |
| itpk1a | 0.382 | ttc36 | -0.221 |
| crip1 | 0.377 | serpina1 | -0.219 |
| GCA | 0.377 | fabp3 | -0.219 |
| eps8l3b | 0.372 | apoa2 | -0.218 |
| tm4sf4 | 0.372 | serpina1l | -0.216 |
| mcl1a | 0.372 | zgc:123103 | -0.214 |
| lxn | 0.372 | rbp2b | -0.213 |
| si:ch1073-443f11.2 | 0.371 | hao1 | -0.212 |
| serpinb1l3 | 0.370 | apom | -0.210 |
| si:dkeyp-72e1.6 | 0.369 | fgg | -0.210 |
| tmsb1 | 0.369 | ppdpfa | -0.208 |
| cldn15la | 0.368 | uox | -0.207 |
| zgc:172079 | 0.366 | fetub | -0.207 |
| calml4a | 0.362 | ambp | -0.207 |
| vil1 | 0.360 | tfa | -0.205 |
| phlda2 | 0.358 | fgb | -0.203 |
| krt92 | 0.357 | fabp10a | -0.203 |
| gsto2 | 0.355 | gatm | -0.202 |
| b2ml | 0.354 | cpn1 | -0.200 |
| fa2h | 0.351 | serpinf2b | -0.200 |
| degs2 | 0.349 | hpda | -0.199 |
| serinc2 | 0.348 | fga | -0.199 |
| lgals2b | 0.348 | vtnb | -0.199 |
| AL831745.1 | 0.346 | ces2 | -0.198 |
| tmem59 | 0.345 | pygl | -0.196 |
| si:dkeyp-73b11.8 | 0.344 | cfhl4 | -0.195 |
| actb1 | 0.343 | LOC110437731 | -0.194 |
| arpc3 | 0.342 | f2 | -0.194 |