myeloid ecotropic viral integration site 3
ZFIN 
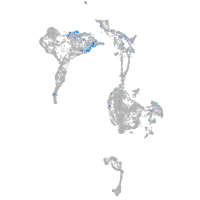
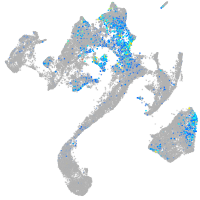
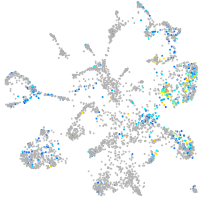
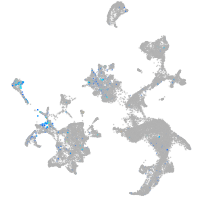
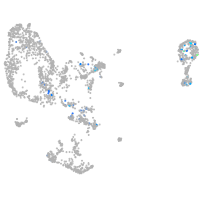
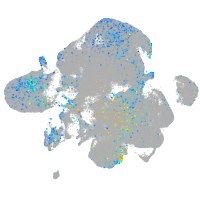
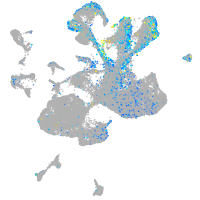
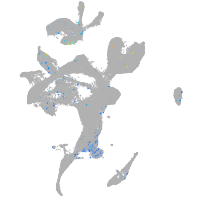
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-016165 | 0.302 | krt8 | -0.075 |
| XLOC-028010 | 0.219 | krt18a.1 | -0.069 |
| BX511073.1 | 0.213 | gas2b | -0.067 |
| pcsk1nl | 0.211 | slc38a5b | -0.064 |
| si:ch211-168d23.3 | 0.197 | atp1b1a | -0.061 |
| BX004991.1 | 0.193 | ucmab | -0.059 |
| map1b | 0.189 | sparc | -0.059 |
| onecut3b | 0.184 | eef1da | -0.059 |
| KCNN2 | 0.178 | efna1b | -0.058 |
| cx39.9 | 0.177 | rgcc | -0.058 |
| lgi3 | 0.177 | mtus1b | -0.057 |
| LOC103909053 | 0.177 | zgc:113232 | -0.055 |
| il17a/f2 | 0.176 | matn3a | -0.055 |
| LOC110438054 | 0.176 | col9a2 | -0.054 |
| spock1 | 0.175 | krt5 | -0.054 |
| GNAZ | 0.175 | npc2 | -0.053 |
| tnfrsf13b | 0.174 | wfdc2 | -0.053 |
| LOC100537384 | 0.172 | si:dkey-31g6.6 | -0.053 |
| slc4a8 | 0.168 | zmp:0000000760 | -0.052 |
| gabra3 | 0.166 | si:ch211-243g18.2 | -0.052 |
| lhx4 | 0.165 | epcam | -0.052 |
| GRIK2 | 0.164 | igsf10 | -0.051 |
| LOC101882015 | 0.160 | col11a2 | -0.051 |
| BX088583.2 | 0.159 | tent5ba | -0.051 |
| syt2a | 0.158 | chad | -0.050 |
| hivep2b | 0.158 | tbx16 | -0.050 |
| urp1 | 0.158 | crabp2a | -0.049 |
| ank3a | 0.157 | epyc | -0.049 |
| snap25a | 0.156 | mmp23bb | -0.049 |
| onecut1 | 0.152 | krt94 | -0.048 |
| gnrhr1 | 0.150 | slc7a8a | -0.048 |
| stxbp1a | 0.150 | cdca7a | -0.048 |
| kcnq2b | 0.149 | pleca | -0.048 |
| drd2a | 0.149 | id3 | -0.048 |
| scn8ab | 0.149 | pitx2 | -0.048 |