lysyl oxidase-like 5a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm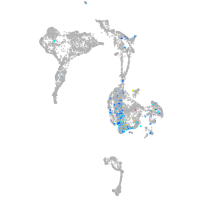 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 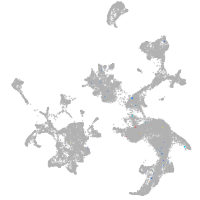 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous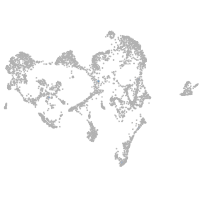 |
pigment cells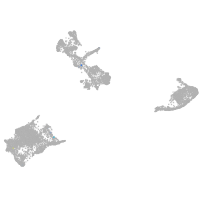 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-37m8.11 | 0.606 | ywhabl | -0.019 |
| dnase1l1l | 0.537 | dynlrb1 | -0.017 |
| mespba | 0.537 | qdpra | -0.017 |
| zgc:173915 | 0.537 | atp6v0ca | -0.017 |
| pgfb | 0.514 | mt-atp6 | -0.017 |
| ACOT12 | 0.422 | ndufb9 | -0.016 |
| LOC110438943 | 0.396 | atp11a | -0.016 |
| tbx22 | 0.366 | gapdhs | -0.016 |
| CR388378.1 | 0.352 | calm2a | -0.016 |
| ccl38.1 | 0.334 | dct | -0.016 |
| slc6a22.2 | 0.305 | tyrp1b | -0.016 |
| CR932000.1 | 0.289 | agtrap | -0.016 |
| LOC101882264 | 0.286 | gpx4b | -0.016 |
| tmem173 | 0.278 | LEPROTL1 | -0.016 |
| si:ch73-334d15.4 | 0.277 | npc2 | -0.015 |
| lrrc15 | 0.275 | tyr | -0.015 |
| sema4aa | 0.264 | mtbl | -0.015 |
| col8a1b | 0.254 | eif4a1a | -0.014 |
| trpc6a | 0.251 | anxa2a | -0.014 |
| gata1a | 0.237 | pmela | -0.014 |
| c1qtnf2 | 0.235 | atp6v1g1 | -0.014 |
| c3b.1 | 0.223 | tyrp1a | -0.014 |
| rflna | 0.219 | zgc:110789 | -0.014 |
| adamts16 | 0.203 | atp5f1d | -0.014 |
| si:ch211-243a20.4 | 0.201 | cotl1 | -0.014 |
| ecel1 | 0.197 | tpm3 | -0.014 |
| si:dkey-15j16.3 | 0.187 | sypl2b | -0.014 |
| kcns3a | 0.185 | cnn3a | -0.014 |
| si:dkey-1b17.1 | 0.174 | phlda1 | -0.014 |
| nt5e | 0.171 | si:ch73-389b16.1 | -0.013 |
| zgc:152753 | 0.170 | ap3d1 | -0.013 |
| fars2 | 0.166 | slc37a2 | -0.013 |
| slc29a1b | 0.165 | hbbe1.2 | -0.013 |
| selenol | 0.162 | fundc1 | -0.013 |
| enpp2 | 0.160 | psmd4a | -0.013 |