lens intrinsic membrane protein 2.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye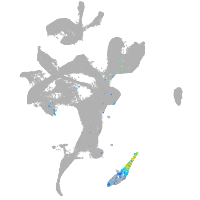 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm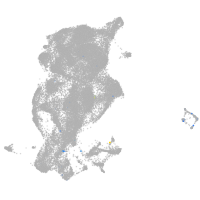 |
fin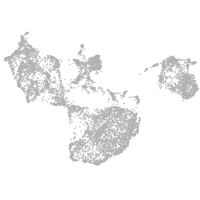 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mipb | 0.469 | rps10 | -0.046 |
| gja8b | 0.461 | rpl37 | -0.045 |
| crygmx | 0.404 | zgc:114188 | -0.043 |
| btg4 | 0.389 | rplp1 | -0.042 |
| lim2.4 | 0.367 | rpl19 | -0.039 |
| cx23 | 0.352 | rpl9 | -0.038 |
| mipa | 0.348 | rplp0 | -0.037 |
| gja8a | 0.346 | rps20 | -0.036 |
| sall1b | 0.337 | nme2b.1 | -0.036 |
| prox2 | 0.337 | rps17 | -0.036 |
| crybb1l1 | 0.315 | prdx2 | -0.036 |
| cryba1b | 0.303 | atp5l | -0.034 |
| si:ch211-255g12.6 | 0.299 | rps3a | -0.034 |
| CT573303.1 | 0.290 | rpl26 | -0.033 |
| lim2.1 | 0.280 | rpl34 | -0.032 |
| si:rp71-45g20.4 | 0.273 | faua | -0.032 |
| crybb1l2 | 0.270 | atp5meb | -0.032 |
| cryba4 | 0.260 | dbi | -0.032 |
| crygn2 | 0.259 | atp5pd | -0.032 |
| pimr179 | 0.248 | ndufb10 | -0.032 |
| LOC101884482 | 0.245 | rpl39 | -0.031 |
| nrl | 0.244 | rpl10 | -0.031 |
| cryba1l1 | 0.241 | eno3 | -0.031 |
| crygm2e | 0.220 | COX7A2 (1 of many) | -0.031 |
| LOC110439554 | 0.216 | suclg1 | -0.031 |
| nrcamb | 0.215 | atp5pb | -0.030 |
| LOC110438002 | 0.209 | ndufb3 | -0.029 |
| nags | 0.207 | zgc:158463 | -0.029 |
| BX000696.1 | 0.207 | cox6a1 | -0.028 |
| fshr | 0.203 | rpl24 | -0.028 |
| s1pr3a | 0.202 | gapdh | -0.028 |
| col28a1b | 0.201 | gsta.1 | -0.028 |
| grifin | 0.200 | ahcy | -0.028 |
| CT583650.1 | 0.196 | rps9 | -0.028 |
| XLOC-025689 | 0.194 | rps14 | -0.028 |