LIM homeobox 9
ZFIN 
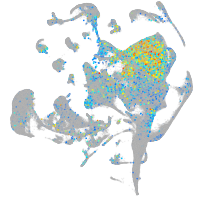
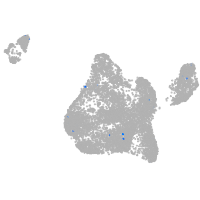
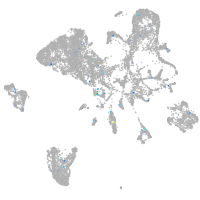
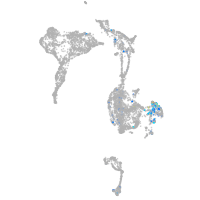
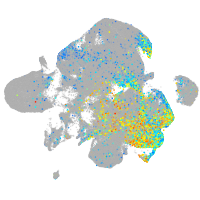
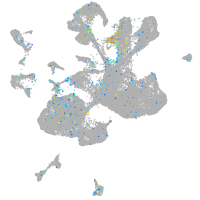
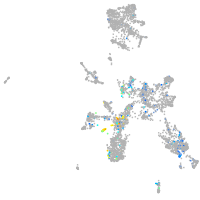
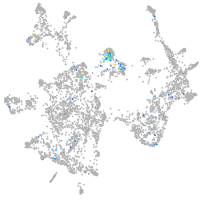
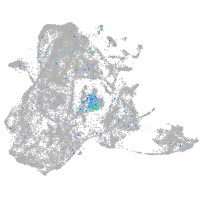
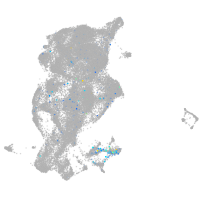
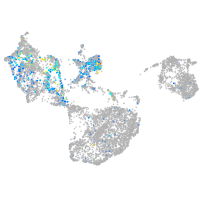
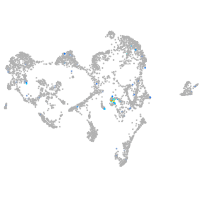
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| barhl2 | 0.366 | smim29 | -0.098 |
| lhx2b | 0.355 | tspan36 | -0.097 |
| elavl3 | 0.338 | rab32a | -0.091 |
| stmn1b | 0.310 | syngr1a | -0.089 |
| adcyap1b | 0.305 | cst14a.2 | -0.088 |
| mab21l1 | 0.293 | sdc4 | -0.087 |
| gpm6aa | 0.292 | trpm1b | -0.085 |
| fez1 | 0.292 | rnaseka | -0.083 |
| ebf3a | 0.289 | gpr143 | -0.083 |
| nova2 | 0.282 | si:dkey-21a6.5 | -0.081 |
| tuba1c | 0.281 | selenoh | -0.081 |
| zc4h2 | 0.275 | pnp4a | -0.078 |
| barhl1a | 0.271 | pcbd1 | -0.078 |
| nrn1a | 0.267 | paics | -0.078 |
| mllt11 | 0.266 | zgc:110239 | -0.077 |
| ptmaa | 0.262 | LOC103910009 | -0.077 |
| tmsb | 0.261 | zgc:110591 | -0.077 |
| tubb5 | 0.261 | slc45a2 | -0.077 |
| gng2 | 0.260 | pttg1ipb | -0.076 |
| pbx3b | 0.260 | si:dkey-151g10.6 | -0.076 |
| rtn1a | 0.255 | impdh1b | -0.075 |
| gng3 | 0.255 | gmps | -0.074 |
| lmo3 | 0.250 | qdpra | -0.074 |
| barhl1b | 0.246 | rab38 | -0.073 |
| bcl11ba | 0.246 | atic | -0.073 |
| pou4f1 | 0.245 | slc38a5b | -0.073 |
| ppp1r14ba | 0.243 | CABZ01048402.2 | -0.072 |
| zgc:65894 | 0.239 | atox1 | -0.072 |
| fyna | 0.238 | slc3a2a | -0.072 |
| hmgb3a | 0.236 | agtrap | -0.072 |
| cbfa2t3 | 0.234 | gstp1 | -0.071 |
| ebf1a | 0.233 | si:dkey-204f11.64 | -0.071 |
| epb41a | 0.233 | rab34b | -0.071 |
| cacnb4b | 0.232 | krt18b | -0.071 |
| hmgb1b | 0.232 | tuba8l | -0.071 |