LIM homeobox 4
ZFIN 
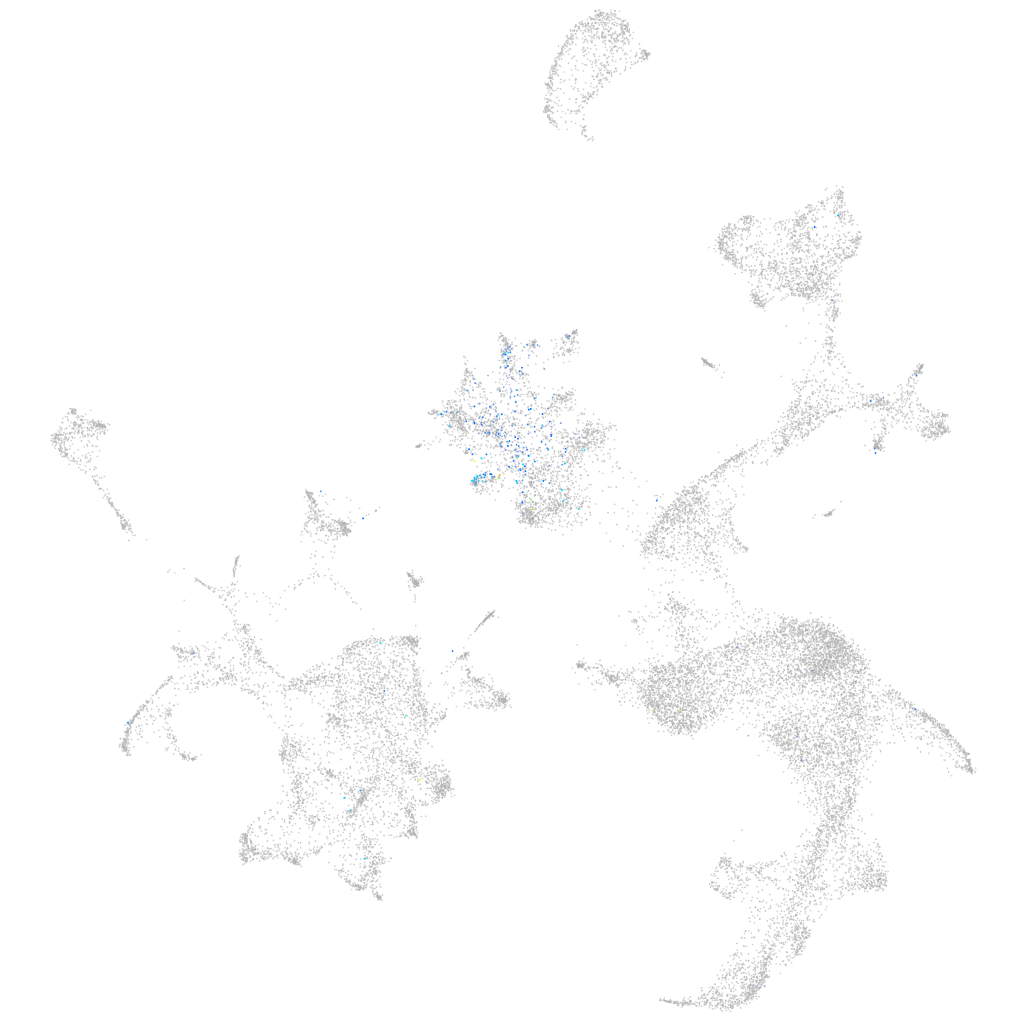
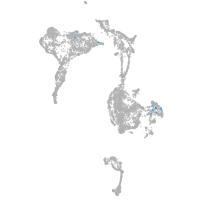
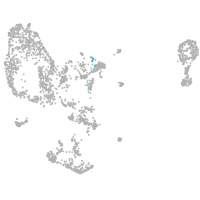
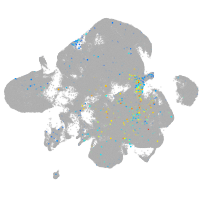
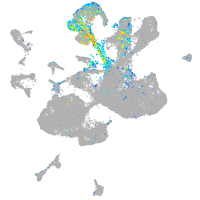
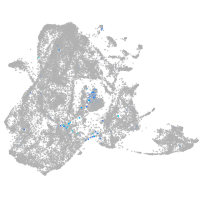
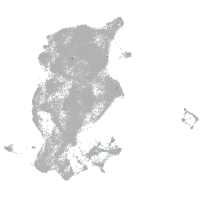
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vsx1 | 0.287 | hbbe3 | -0.033 |
| syt5b | 0.285 | pfn1 | -0.033 |
| crx | 0.253 | arl4aa | -0.030 |
| otx5 | 0.250 | hhex | -0.030 |
| vamp1 | 0.244 | mafbb | -0.030 |
| pcp4l1 | 0.233 | hmbsa | -0.029 |
| neurod4 | 0.231 | rab11fip1b | -0.027 |
| sypb | 0.224 | dusp5 | -0.027 |
| nrn1lb | 0.222 | gmfg | -0.027 |
| epb41a | 0.222 | znfl2a | -0.027 |
| snap25b | 0.222 | laptm5 | -0.026 |
| cplx4a | 0.221 | arhgdia | -0.026 |
| foxg1b | 0.220 | fli1b | -0.026 |
| rs1a | 0.217 | rac2 | -0.026 |
| ppp1r1b | 0.216 | CT030188.1 | -0.026 |
| calb2b | 0.216 | clic1 | -0.025 |
| cntn1b | 0.215 | ctss2.1 | -0.025 |
| dscaml1 | 0.213 | wasb | -0.025 |
| eml1 | 0.210 | XLOC-014079 | -0.025 |
| si:ch73-256g18.2 | 0.209 | alad | -0.025 |
| bhlhe23 | 0.209 | clec14a | -0.025 |
| neurod1 | 0.208 | lmo2 | -0.024 |
| gng13b | 0.207 | srgn | -0.024 |
| lin7a | 0.205 | gsto2 | -0.024 |
| ndrg4 | 0.204 | capgb | -0.024 |
| gnb3a | 0.203 | inpp5d | -0.024 |
| nsg2 | 0.201 | cldng | -0.024 |
| golga7ba | 0.200 | myct1a | -0.024 |
| mpp6b | 0.197 | gnpda1 | -0.024 |
| scg3 | 0.196 | sptlc2a | -0.024 |
| gpm6aa | 0.196 | spi1b | -0.024 |
| six3b | 0.196 | si:ch73-248e21.7 | -0.024 |
| atp6v0cb | 0.195 | etv2 | -0.024 |
| cnrip1b | 0.193 | coro1a | -0.023 |
| irx6a | 0.191 | glrx5 | -0.023 |