lefty1
ZFIN 
Other cell groups


all cells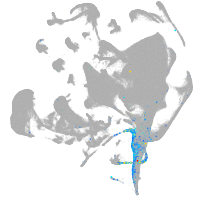 |
blastomeres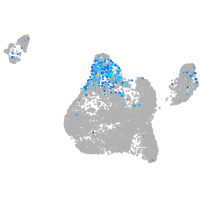 |
PGCs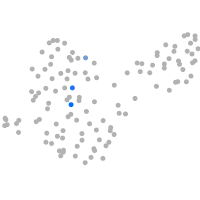 |
endoderm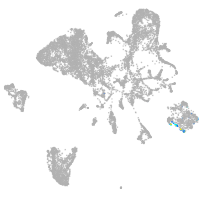 |
axial mesoderm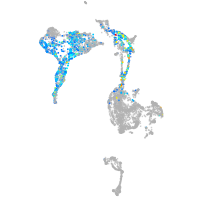 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 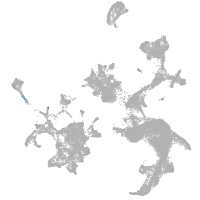 |
pronephros 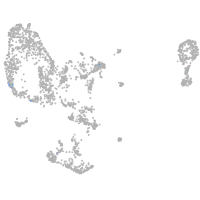 |
neural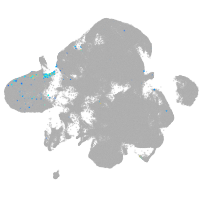 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lft2 | 0.636 | rpl37 | -0.103 |
| ndr2 | 0.533 | rps10 | -0.103 |
| nkx2.9 | 0.469 | zgc:114188 | -0.091 |
| nkx2.4b | 0.432 | h3f3a | -0.082 |
| gsc | 0.428 | CR383676.1 | -0.076 |
| nkx2.2b | 0.348 | hnrnpa0l | -0.072 |
| dbx1a | 0.330 | rps17 | -0.072 |
| dact2 | 0.328 | ppiab | -0.071 |
| foxa | 0.294 | ptmaa | -0.070 |
| nkx1.2la | 0.285 | si:ch1073-429i10.3.1 | -0.060 |
| foxb1b | 0.277 | zgc:158463 | -0.058 |
| nkx2.1 | 0.264 | mt-atp6 | -0.055 |
| tdgf1 | 0.232 | mt-nd1 | -0.055 |
| admp | 0.229 | COX3 | -0.053 |
| foxa3 | 0.228 | cct2 | -0.050 |
| foxa2 | 0.226 | atp5f1b | -0.050 |
| foxd5 | 0.225 | marcksl1a | -0.050 |
| sox19a | 0.209 | fabp3 | -0.048 |
| shisa2a | 0.207 | ckbb | -0.047 |
| fgf3 | 0.207 | h3f3c | -0.046 |
| shha | 0.206 | gpm6aa | -0.046 |
| fzd8b | 0.196 | tuba1c | -0.045 |
| nkx2.4a | 0.194 | fabp7a | -0.045 |
| hspb1 | 0.192 | zgc:153409 | -0.045 |
| stm | 0.186 | atp5mc1 | -0.043 |
| pitx2 | 0.177 | cadm3 | -0.042 |
| nr6a1a | 0.170 | zgc:56493 | -0.042 |
| COX7A2 | 0.167 | mt-co2 | -0.041 |
| ISCU | 0.164 | gpm6ab | -0.039 |
| si:ch211-152c2.3 | 0.164 | gapdhs | -0.039 |
| fzd8a | 0.164 | ppdpfb | -0.038 |
| noto | 0.158 | atp5mc3b | -0.038 |
| XLOC-043201 | 0.152 | NC-002333.17 | -0.037 |
| CR847971.1 | 0.150 | pvalb1 | -0.037 |
| LOC101882159 | 0.150 | celf2 | -0.036 |




