LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
ZFIN 
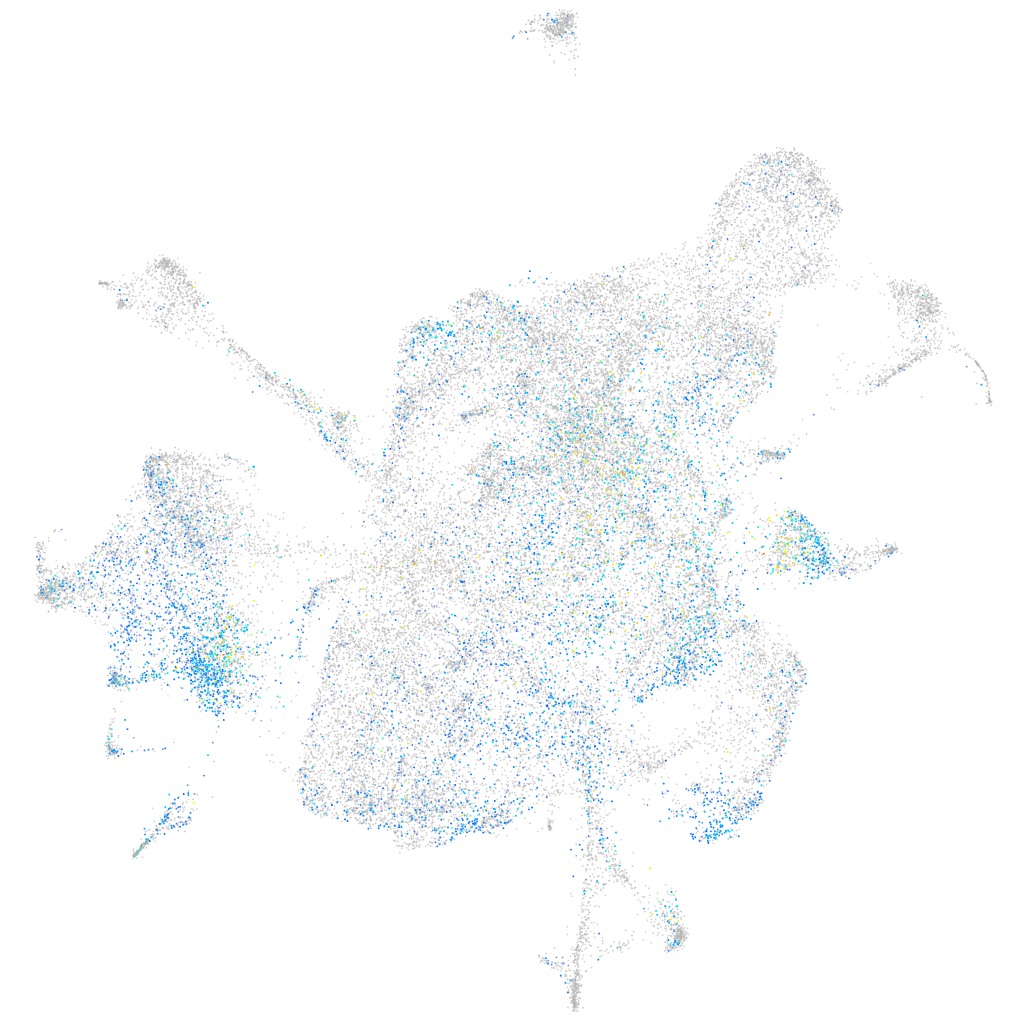
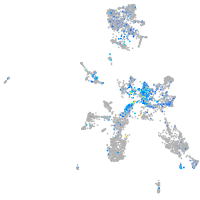
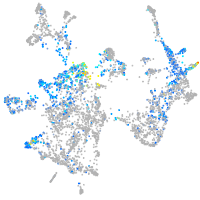
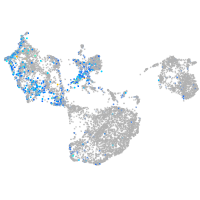
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dla | 0.189 | fgfbp2b | -0.113 |
| her4.2 | 0.186 | col11a2 | -0.083 |
| XLOC-003689 | 0.182 | matn1 | -0.077 |
| her4.1 | 0.177 | col2a1a | -0.074 |
| XLOC-003692 | 0.171 | col9a1a | -0.071 |
| abhd6a | 0.163 | ucmab | -0.071 |
| XLOC-003690 | 0.162 | si:ch211-156j16.1 | -0.071 |
| her15.1 | 0.160 | col9a3 | -0.070 |
| si:ch73-21g5.7 | 0.151 | col9a2 | -0.069 |
| her12 | 0.146 | mia | -0.069 |
| LOC798783 | 0.140 | acana | -0.067 |
| sox3 | 0.138 | si:dkey-19b23.8 | -0.067 |
| sox19a | 0.138 | wwp2 | -0.067 |
| dld | 0.130 | epyc | -0.067 |
| marcksl1a | 0.125 | snorc | -0.067 |
| her4.4 | 0.123 | sox9a | -0.064 |
| CU467822.1 | 0.123 | wisp3 | -0.063 |
| CU634008.1 | 0.123 | col11a1a | -0.062 |
| gadd45gb.1 | 0.122 | tnxba | -0.059 |
| notch1a | 0.119 | rock2b | -0.058 |
| dlb | 0.116 | gyg1b | -0.056 |
| her2 | 0.113 | ptgdsa | -0.056 |
| zgc:165461 | 0.113 | VIT | -0.055 |
| marcksl1b | 0.109 | fgfr3 | -0.055 |
| thbs4b | 0.108 | pcolceb | -0.054 |
| cilp | 0.099 | otos | -0.054 |
| notch3 | 0.098 | emilin3a | -0.054 |
| pax3a | 0.097 | si:dkey-6n6.1 | -0.054 |
| CR848047.1 | 0.097 | klf2b | -0.054 |
| her13 | 0.095 | foxc1b | -0.054 |
| lrrn1 | 0.094 | hapln1b | -0.053 |
| gpm6bb | 0.094 | barx1 | -0.053 |
| fabp3 | 0.093 | anxa1a | -0.053 |
| plp1a | 0.093 | col8a2 | -0.053 |
| notch1b | 0.092 | AL929222.2 | -0.052 |