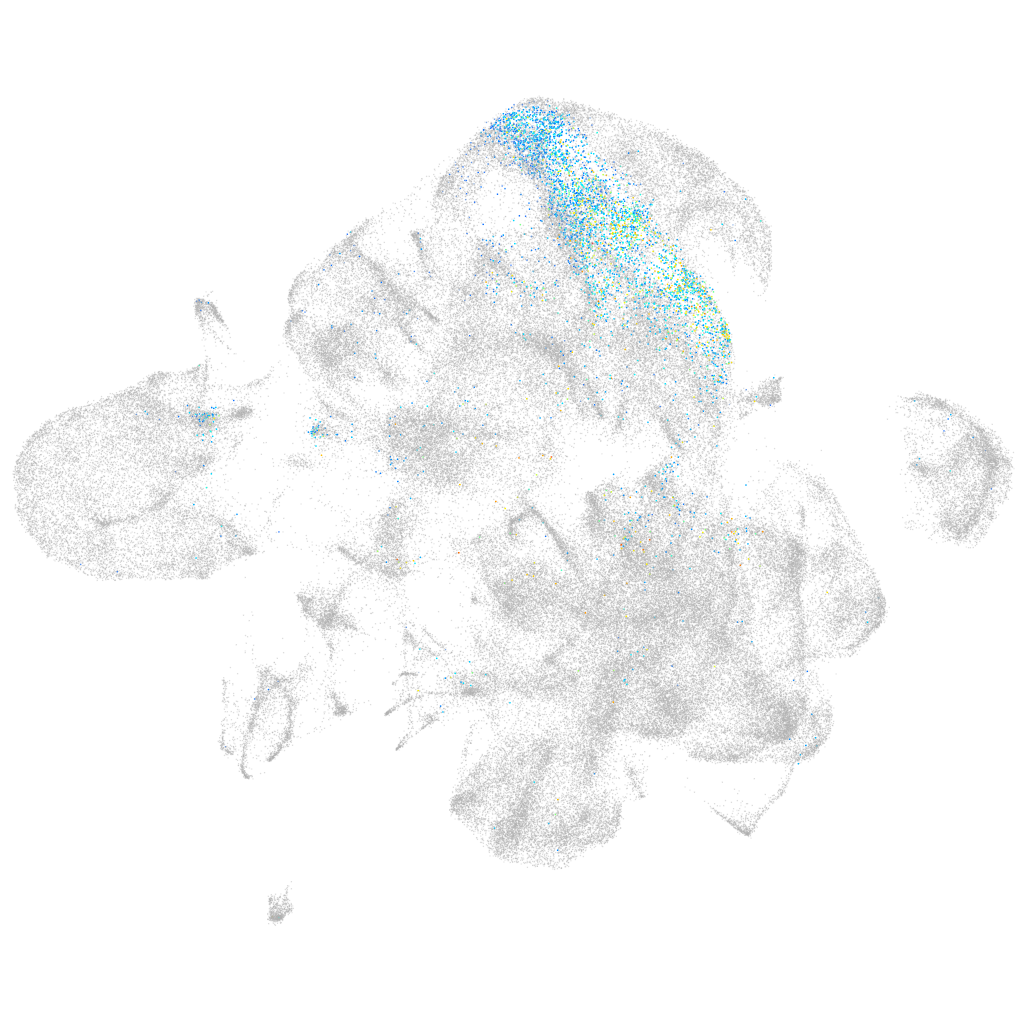
ladybird homeobox 1b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gsx1 | 0.316 | stmn1b | -0.095 |
| AL845362.1 | 0.302 | gng3 | -0.094 |
| pax3a | 0.277 | gpm6ab | -0.093 |
| prdm13 | 0.235 | sncb | -0.091 |
| prss35 | 0.230 | ckbb | -0.086 |
| gas1b | 0.209 | rtn1b | -0.085 |
| sox21a | 0.206 | atp6v0cb | -0.083 |
| mdka | 0.202 | elavl4 | -0.081 |
| slc1a3a | 0.199 | vamp2 | -0.080 |
| egfl6 | 0.199 | ywhag2 | -0.079 |
| hs3st3b1a | 0.196 | stmn2a | -0.079 |
| gfap | 0.190 | gapdhs | -0.078 |
| plp1a | 0.185 | stx1b | -0.077 |
| XLOC-003689 | 0.181 | zgc:65894 | -0.075 |
| endouc | 0.181 | gap43 | -0.074 |
| nppc | 0.181 | fez1 | -0.074 |
| lbx1a | 0.180 | atp6v1e1b | -0.073 |
| ptf1a | 0.179 | mllt11 | -0.072 |
| XLOC-003690 | 0.179 | tmsb2 | -0.072 |
| si:ch211-286o17.1 | 0.178 | snap25a | -0.072 |
| sb:cb81 | 0.177 | stxbp1a | -0.070 |
| gas1a | 0.176 | atpv0e2 | -0.070 |
| boc | 0.168 | olfm1b | -0.069 |
| si:dkey-188g12.1 | 0.166 | calm1a | -0.069 |
| kirrel3l | 0.165 | map1aa | -0.067 |
| sox3 | 0.164 | id4 | -0.067 |
| cldn5a | 0.162 | rnasekb | -0.067 |
| auts2b | 0.160 | csdc2a | -0.066 |
| CABZ01075068.1 | 0.155 | aplp1 | -0.066 |
| lrrn1 | 0.151 | atp6v1g1 | -0.066 |
| cyp26b1 | 0.150 | h2afy2 | -0.066 |
| notch3 | 0.150 | atp1a3a | -0.065 |
| angptl4 | 0.148 | tmem59l | -0.065 |
| hoxa2b | 0.147 | calm2a | -0.065 |
| zbtb16a | 0.147 | si:dkeyp-75h12.5 | -0.064 |