Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 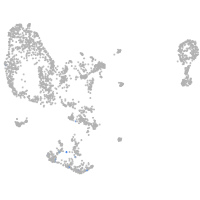 |
neural |
spinal cord / glia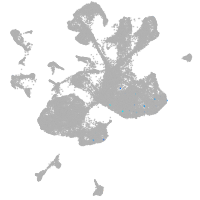 |
eye |
taste / olfactory |
otic / lateral line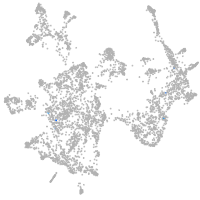 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-208k4.2 | 0.082 | gpm6aa | -0.017 |
| dazl | 0.078 | gpm6ab | -0.017 |
| wee2 | 0.077 | hbae3 | -0.017 |
| tdrd7a | 0.076 | hbbe1.3 | -0.017 |
| btg4 | 0.074 | pvalb1 | -0.017 |
| zgc:173742 | 0.074 | pvalb2 | -0.017 |
| dnd1 | 0.073 | stmn1b | -0.017 |
| zgc:113424 | 0.073 | atp6v0cb | -0.016 |
| ca15b | 0.072 | rtn1a | -0.016 |
| cldnd | 0.072 | actc1b | -0.015 |
| buc | 0.071 | ccni | -0.015 |
| pabpn1l | 0.071 | elavl3 | -0.015 |
| si:rp71-45k5.2 | 0.071 | gapdhs | -0.015 |
| sinup | 0.071 | gng3 | -0.015 |
| eloal | 0.070 | hbae1.1 | -0.015 |
| gtf3ab | 0.070 | hbbe1.1 | -0.015 |
| h1m | 0.070 | calm1a | -0.015 |
| buc2l | 0.069 | sncb | -0.014 |
| cdca9 | 0.069 | tuba1c | -0.014 |
| nanog | 0.069 | vamp2 | -0.014 |
| lsm14b | 0.068 | marcksl1a | -0.014 |
| map1lc3c | 0.067 | ckbb | -0.013 |
| acp5a | 0.066 | rnasekb | -0.013 |
| fam113 | 0.066 | rtn1b | -0.013 |
| ccna1 | 0.065 | wu:fb55g09 | -0.013 |
| si:ch211-145c1.1 | 0.065 | atp6v1e1b | -0.012 |
| h2af1al | 0.064 | cyt1 | -0.012 |
| tatdn2 | 0.064 | elavl4 | -0.012 |
| bmp15 | 0.063 | fez1 | -0.012 |
| pabpc1l | 0.063 | hbbe1.2 | -0.012 |
| cth1 | 0.062 | ip6k2a | -0.012 |
| larp6b | 0.062 | kdm6bb | -0.012 |
| rbm46 | 0.062 | krt4 | -0.012 |
| tmem144b | 0.062 | mylpfa | -0.012 |
| mcm3l | 0.061 | myt1b | -0.012 |




