deleted in azoospermia-like
ZFIN 
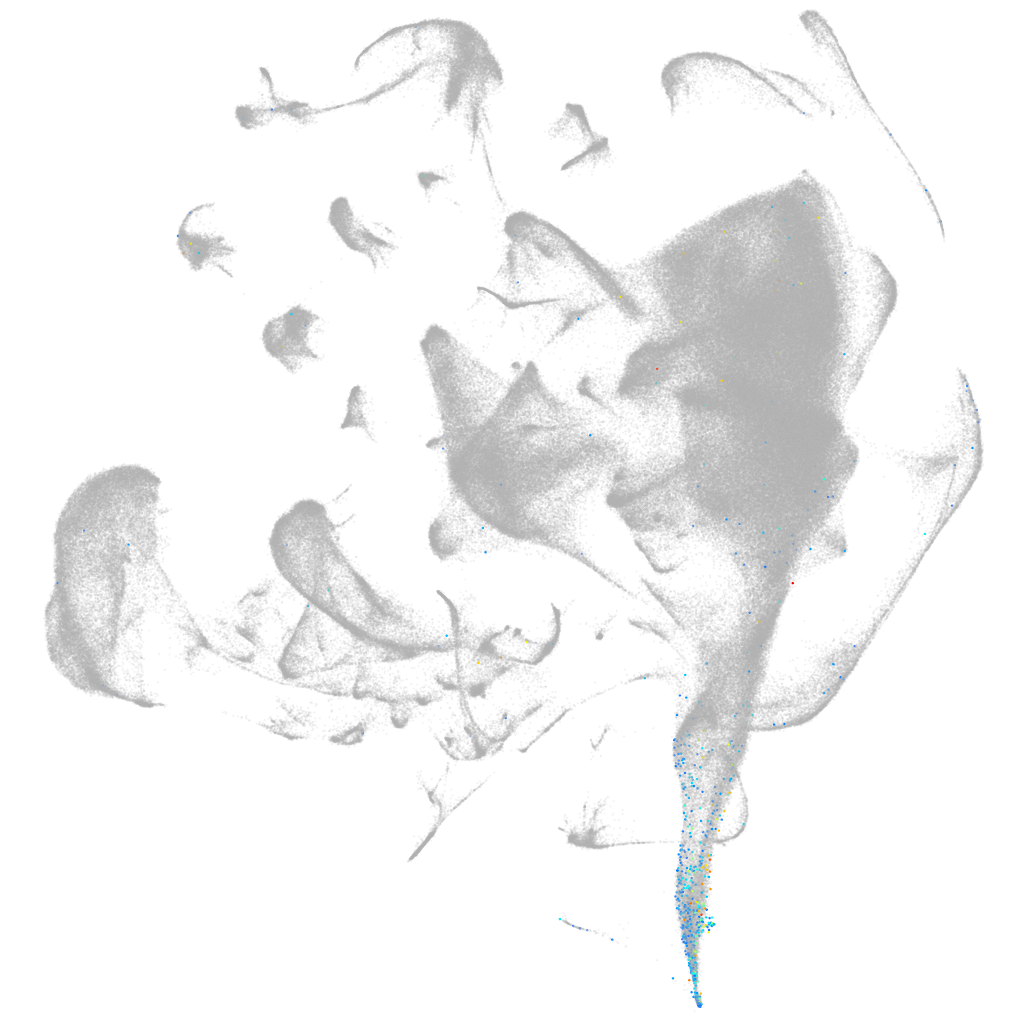
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dnd1 | 0.217 | calm1a | -0.039 |
| tdrd7a | 0.210 | gpm6aa | -0.038 |
| h1m | 0.209 | tuba1c | -0.038 |
| ca15b | 0.208 | atp6v0cb | -0.037 |
| si:dkey-208k4.2 | 0.202 | gapdhs | -0.037 |
| ddx4 | 0.201 | hbbe1.3 | -0.037 |
| zgc:173742 | 0.196 | ccni | -0.036 |
| pabpn1l | 0.192 | hbae3 | -0.036 |
| nanog | 0.191 | rnasekb | -0.036 |
| zgc:113424 | 0.189 | rtn1a | -0.036 |
| ccna1 | 0.182 | stmn1b | -0.035 |
| cldnd | 0.180 | si:ch1073-429i10.3.1 | -0.035 |
| avd | 0.178 | gng3 | -0.034 |
| wee2 | 0.178 | nova2 | -0.034 |
| cth1 | 0.177 | gpm6ab | -0.033 |
| si:rp71-45k5.2 | 0.177 | hbae1.1 | -0.033 |
| h2af1al | 0.174 | krt4 | -0.032 |
| acp5a | 0.170 | pvalb1 | -0.032 |
| zgc:77118 | 0.168 | vamp2 | -0.032 |
| cyp11a1 | 0.167 | atp6v1e1b | -0.031 |
| map1lc3c | 0.166 | elavl3 | -0.031 |
| gra | 0.165 | hbbe1.1 | -0.031 |
| larp6b | 0.164 | mdkb | -0.031 |
| btg4 | 0.163 | cyt1 | -0.030 |
| FO834825.1 | 0.162 | pvalb2 | -0.030 |
| si:dkey-68o6.5 | 0.162 | sncb | -0.030 |
| tatdn2 | 0.161 | marcksl1a | -0.030 |
| zgc:103482 | 0.161 | elavl4 | -0.029 |
| fam113 | 0.160 | olfm1b | -0.029 |
| gtf3ab | 0.158 | snap25a | -0.029 |
| lsm14b | 0.158 | stx1b | -0.029 |
| nanos3 | 0.158 | wu:fb55g09 | -0.029 |
| gdf3 | 0.157 | ywhag2 | -0.029 |
| pabpc1l | 0.157 | zgc:65894 | -0.029 |
| sinup | 0.157 | stxbp1a | -0.029 |