Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 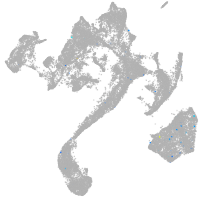 |
mural cells / non-skeletal muscle  |
mesenchyme 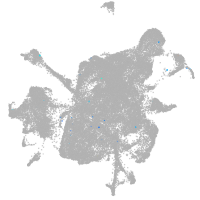 |
hematopoietic / vasculature 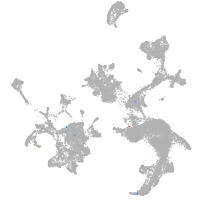 |
pronephros 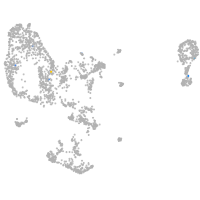 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.026 | gpm6aa | -0.014 |
| dkc1 | 0.024 | tuba1c | -0.014 |
| fbl | 0.024 | ckbb | -0.013 |
| nop58 | 0.024 | rtn1a | -0.013 |
| bms1 | 0.023 | marcksl1a | -0.013 |
| ncl | 0.023 | elavl3 | -0.012 |
| nhp2 | 0.023 | gpm6ab | -0.012 |
| nop2 | 0.023 | pvalb1 | -0.012 |
| nop56 | 0.023 | pvalb2 | -0.012 |
| eif5a2 | 0.022 | stmn1b | -0.012 |
| gnl3 | 0.022 | atp6v0cb | -0.011 |
| mybbp1a | 0.022 | ccni | -0.011 |
| rsl1d1 | 0.022 | gng3 | -0.011 |
| snu13b | 0.022 | hbbe1.3 | -0.011 |
| banf1 | 0.021 | nova2 | -0.011 |
| ddx18 | 0.021 | rnasekb | -0.011 |
| ebna1bp2 | 0.021 | atp6v1g1 | -0.010 |
| eif4bb | 0.021 | epb41a | -0.010 |
| lyar | 0.021 | hbae1.1 | -0.010 |
| mphosph10 | 0.021 | hbae3 | -0.010 |
| nolc1 | 0.021 | myt1b | -0.010 |
| pes | 0.021 | sncb | -0.010 |
| rrp1 | 0.021 | tmsb | -0.010 |
| chaf1a | 0.020 | vamp2 | -0.010 |
| ddx27 | 0.020 | calm1a | -0.010 |
| gar1 | 0.020 | actc1b | -0.009 |
| hspb1 | 0.020 | atp6v1e1b | -0.009 |
| kri1 | 0.020 | atpv0e2 | -0.009 |
| lig1 | 0.020 | cspg5a | -0.009 |
| nop10 | 0.020 | CU467822.1 | -0.009 |
| pprc1 | 0.020 | dpysl3 | -0.009 |
| si:ch211-217k17.7 | 0.020 | elavl4 | -0.009 |
| UTP14C | 0.020 | fez1 | -0.009 |
| wdr43 | 0.020 | gapdhs | -0.009 |
| zmp:0000000624 | 0.020 | gng2 | -0.009 |




