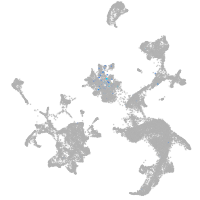
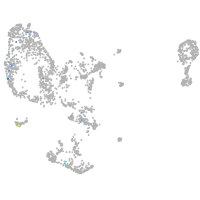
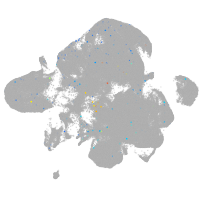
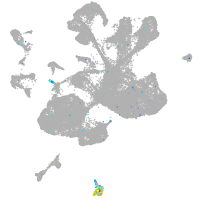
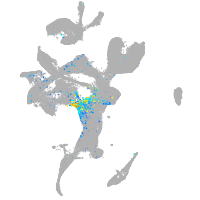
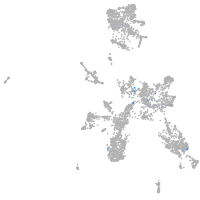
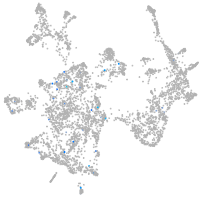
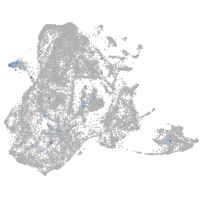
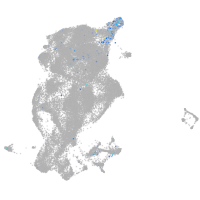
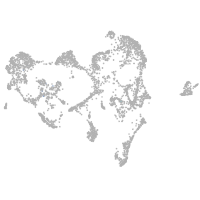
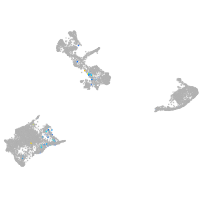
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcnk10a | 0.361 | rps8a | -0.039 |
| rrh | 0.347 | eno3 | -0.039 |
| togaram1 | 0.325 | sec61b | -0.037 |
| LOC103909937 | 0.303 | ppib | -0.036 |
| XLOC-042515 | 0.284 | abat | -0.036 |
| si:dkey-27b3.4 | 0.271 | ahcy | -0.035 |
| si:ch211-264f5.8 | 0.256 | atp5pd | -0.035 |
| LO017650.1 | 0.255 | ssr4 | -0.035 |
| CABZ01077555.1 | 0.251 | cox5ab | -0.033 |
| CU929418.2 | 0.247 | si:ch1073-325m22.2 | -0.033 |
| CABZ01087091.1 | 0.241 | mat1a | -0.032 |
| zgc:113274 | 0.231 | chchd10 | -0.032 |
| hyal6 | 0.231 | kng1 | -0.032 |
| LOC110438742 | 0.223 | BX908782.3 | -0.032 |
| BX005448.1 | 0.222 | lipf | -0.032 |
| si:rp71-1i20.2 | 0.220 | sec61g | -0.032 |
| CT955996.1 | 0.217 | LOC110437731 | -0.032 |
| LOC103909610 | 0.216 | msrb2 | -0.031 |
| cplx3b | 0.213 | c9 | -0.031 |
| LOC101884506 | 0.210 | ssr2 | -0.031 |
| serinc4 | 0.209 | krtcap2 | -0.031 |
| ca14 | 0.207 | eef1da | -0.031 |
| zgc:92912 | 0.207 | mgst1.2 | -0.031 |
| zgc:165604 | 0.206 | aldob | -0.031 |
| gpr186 | 0.204 | pgk1 | -0.030 |
| thsd7ab | 0.198 | agt | -0.030 |
| LOC101883622 | 0.195 | serpinc1 | -0.030 |
| fgf8b | 0.194 | epdl1 | -0.030 |
| LOC101884740 | 0.190 | hadh | -0.030 |
| LOC108190565 | 0.190 | gapdh | -0.030 |
| rx1 | 0.188 | agxta | -0.030 |
| fam131ba | 0.188 | hsp90b1 | -0.030 |
| rai1 | 0.187 | serpinf2a | -0.030 |
| mfsd2b | 0.185 | pnp4b | -0.030 |
| gpr37a | 0.185 | ces2 | -0.030 |