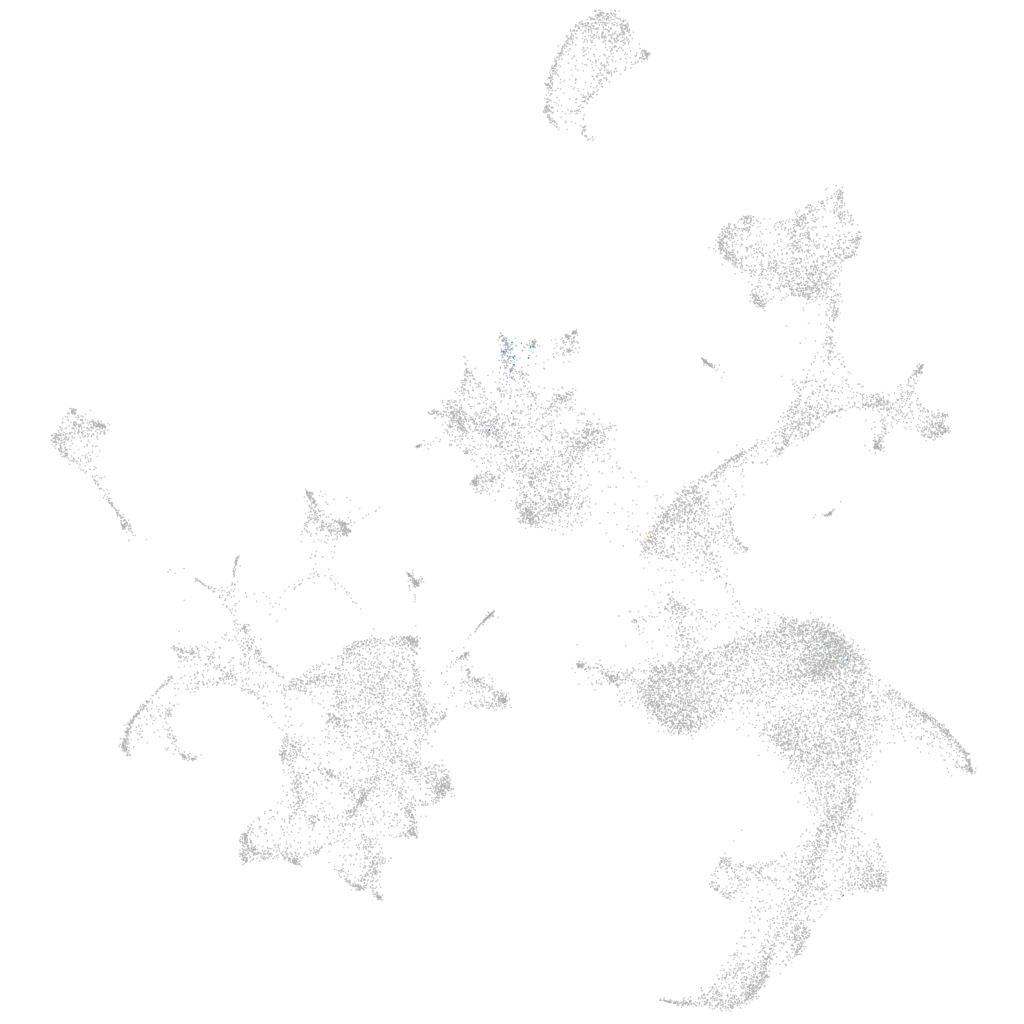
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mir729 | 0.277 | srgn | -0.016 |
| BX649620.1 | 0.276 | fthl27 | -0.015 |
| si:dkey-19c16.12 | 0.258 | zgc:153867 | -0.015 |
| lrit2 | 0.253 | lmo2 | -0.015 |
| zgc:103625 | 0.243 | sat1a.2 | -0.015 |
| gabra6a | 0.240 | tagln2 | -0.015 |
| cngb3.2 | 0.229 | msna | -0.014 |
| arl3l2 | 0.226 | cd63 | -0.014 |
| tulp1b | 0.223 | cebpb | -0.013 |
| cacna1fa | 0.222 | cdh5 | -0.013 |
| cnga3a | 0.219 | rhocb | -0.013 |
| tmx3a | 0.218 | rac2 | -0.013 |
| rgs9a | 0.218 | yrk | -0.013 |
| faimb | 0.215 | ponzr1 | -0.013 |
| elovl4b | 0.214 | clec14a | -0.013 |
| cdhr1a | 0.212 | dusp5 | -0.013 |
| LOC101886132 | 0.209 | arhgdia | -0.013 |
| atp1b2b | 0.202 | krt8 | -0.012 |
| si:dkey-17e16.15 | 0.198 | etv2 | -0.012 |
| ckmt2a | 0.197 | cdc42l | -0.012 |
| arl3l1 | 0.194 | hhex | -0.012 |
| grk1b | 0.194 | coro1a | -0.012 |
| rcvrn2 | 0.193 | CT030188.1 | -0.012 |
| slc25a3a | 0.190 | sox7 | -0.012 |
| rcvrna | 0.189 | sult6b1 | -0.012 |
| guk1b | 0.189 | egfl7 | -0.012 |
| prph2b | 0.189 | gng5 | -0.012 |
| pdcb | 0.188 | mcamb | -0.012 |
| rbp4l | 0.186 | tuba8l | -0.012 |
| grk7a | 0.186 | atp1b1a | -0.012 |
| tmem237a | 0.185 | tpm3 | -0.012 |
| si:dkeyp-50f7.2 | 0.184 | fli1a | -0.012 |
| inaa | 0.183 | csrp1a | -0.012 |
| prph2a | 0.182 | ramp2 | -0.012 |
| si:dkey-220f10.4 | 0.181 | si:ch73-248e21.7 | -0.012 |