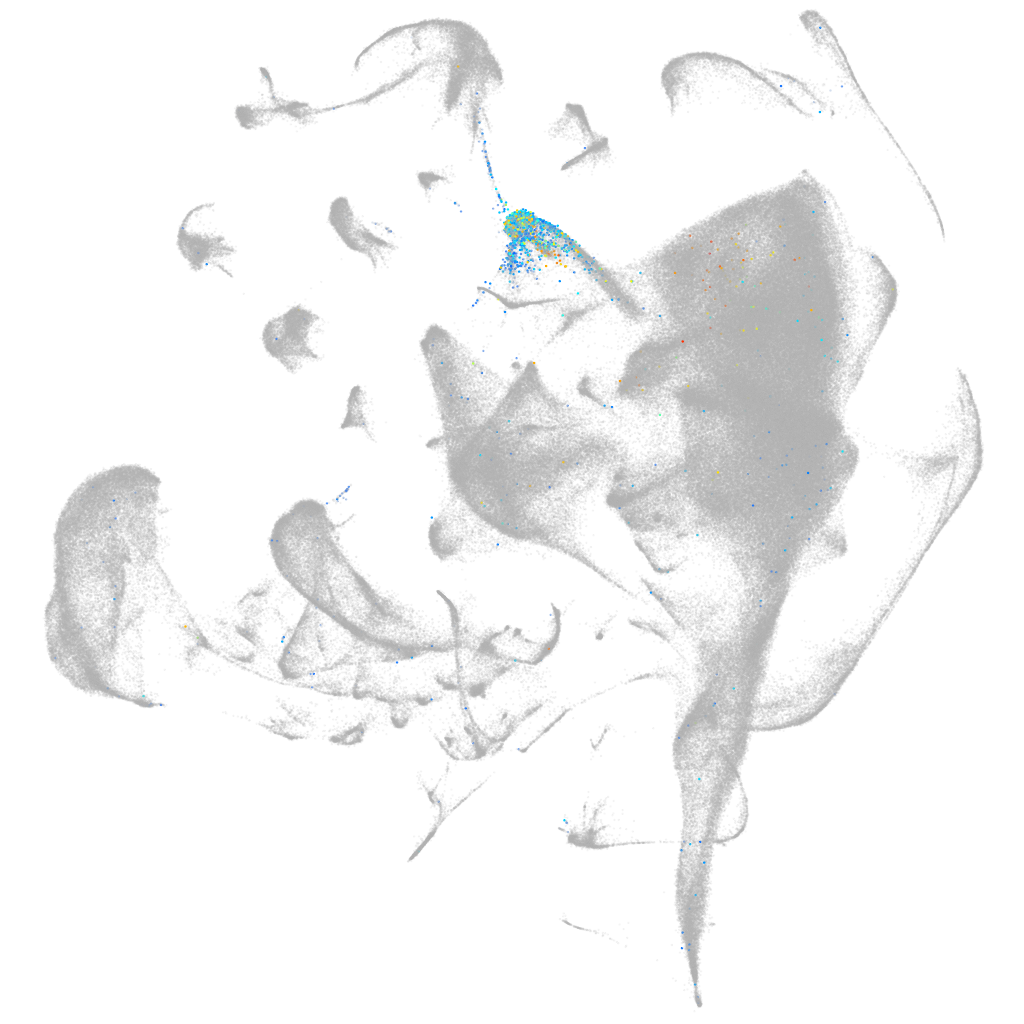
"calcium channel, voltage-dependent, L type, alpha 1F subunit a"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| guk1b | 0.391 | rtn1a | -0.034 |
| slc25a3a | 0.389 | ctsla | -0.032 |
| cnga3a | 0.381 | si:dkey-56m19.5 | -0.032 |
| tmx3a | 0.379 | marcksb | -0.031 |
| ckmt2a | 0.378 | rtn1b | -0.031 |
| si:dkey-17e16.15 | 0.378 | nova2 | -0.029 |
| cngb3.2 | 0.373 | tmeff1b | -0.029 |
| rgs9a | 0.372 | zgc:56493 | -0.029 |
| pdcb | 0.371 | elavl3 | -0.028 |
| rcvrn2 | 0.371 | zc4h2 | -0.028 |
| arl3l2 | 0.369 | marcksl1a | -0.028 |
| rcvrna | 0.364 | atp2b2 | -0.027 |
| prph2a | 0.360 | calm2a | -0.027 |
| elovl4b | 0.358 | cd99l2 | -0.027 |
| faimb | 0.357 | tubb5 | -0.027 |
| gnat2 | 0.356 | fam168a | -0.026 |
| gnb3b | 0.354 | qkia | -0.026 |
| grk7a | 0.352 | si:ch73-46j18.5 | -0.026 |
| arl3l1 | 0.351 | stmn1b | -0.026 |
| si:ch1073-469d17.2 | 0.350 | ywhag2 | -0.026 |
| si:ch211-207l14.1 | 0.350 | btg2 | -0.025 |
| prph2b | 0.347 | ctnnb1 | -0.025 |
| grk1b | 0.345 | elavl4 | -0.025 |
| aipl2 | 0.344 | fxyd6l | -0.025 |
| zgc:73359 | 0.343 | gng3 | -0.025 |
| sv2ba | 0.340 | mdkb | -0.025 |
| tmem237a | 0.340 | si:ch211-288g17.3 | -0.025 |
| tmem136b | 0.338 | stx1b | -0.025 |
| arr3a | 0.335 | tmem59l | -0.025 |
| impg2a | 0.335 | tpm3 | -0.025 |
| zgc:103625 | 0.335 | ywhaz | -0.025 |
| pde6c | 0.332 | stxbp1a | -0.025 |
| si:dkey-220f10.4 | 0.330 | aldob | -0.024 |
| si:ch1073-155h21.2 | 0.329 | aplp1 | -0.024 |
| gngt2b | 0.327 | gng5 | -0.024 |