Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye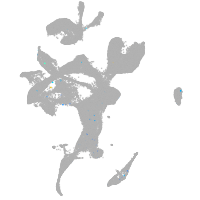 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:zfos-451d2.2 | 0.094 | elavl3 | -0.033 |
| XLOC-008391 | 0.071 | rtn1a | -0.032 |
| zgc:110540 | 0.054 | stmn1b | -0.029 |
| si:ch211-217i17.1 | 0.053 | ckbb | -0.027 |
| pcna | 0.051 | gpm6aa | -0.027 |
| dut | 0.051 | gpm6ab | -0.026 |
| fen1 | 0.050 | tubb5 | -0.024 |
| cdc6 | 0.049 | tmsb | -0.023 |
| selenoh | 0.048 | hbbe1.3 | -0.023 |
| cdkn1a | 0.048 | marcksl1b | -0.023 |
| dhfr | 0.047 | gng3 | -0.022 |
| rrm2 | 0.047 | hmgb3a | -0.022 |
| LO018605.3 | 0.047 | hbae3 | -0.021 |
| rpa2 | 0.047 | tuba1a | -0.021 |
| adgre13 | 0.046 | dpysl3 | -0.021 |
| unga | 0.046 | fez1 | -0.021 |
| rfc4 | 0.045 | rnasekb | -0.021 |
| shmt1 | 0.044 | rtn1b | -0.021 |
| bspry | 0.044 | hbae1.1 | -0.021 |
| prim2 | 0.044 | ip6k2a | -0.020 |
| rrm1 | 0.044 | nova2 | -0.020 |
| her2 | 0.043 | myt1b | -0.020 |
| GALNT3 | 0.043 | si:dkeyp-75h12.5 | -0.020 |
| mcm4 | 0.043 | pvalb2 | -0.020 |
| nutf2l | 0.042 | fscn1a | -0.020 |
| si:ch211-51i16.1 | 0.042 | zc4h2 | -0.020 |
| btg3 | 0.042 | aplp1 | -0.019 |
| si:ch211-171h4.7 | 0.042 | COX3 | -0.019 |
| msh6 | 0.042 | si:dkey-56m19.5 | -0.019 |
| gmnn | 0.042 | elavl4 | -0.019 |
| zgc:56493 | 0.042 | kdm6bb | -0.019 |
| LOC100330864 | 0.042 | ebf3a | -0.019 |
| mcm7 | 0.042 | chd4a | -0.019 |
| rnaseh2a | 0.041 | nsg2 | -0.019 |
| slbp | 0.041 | csdc2a | -0.019 |




