kelch repeat-containing protein
ZFIN 
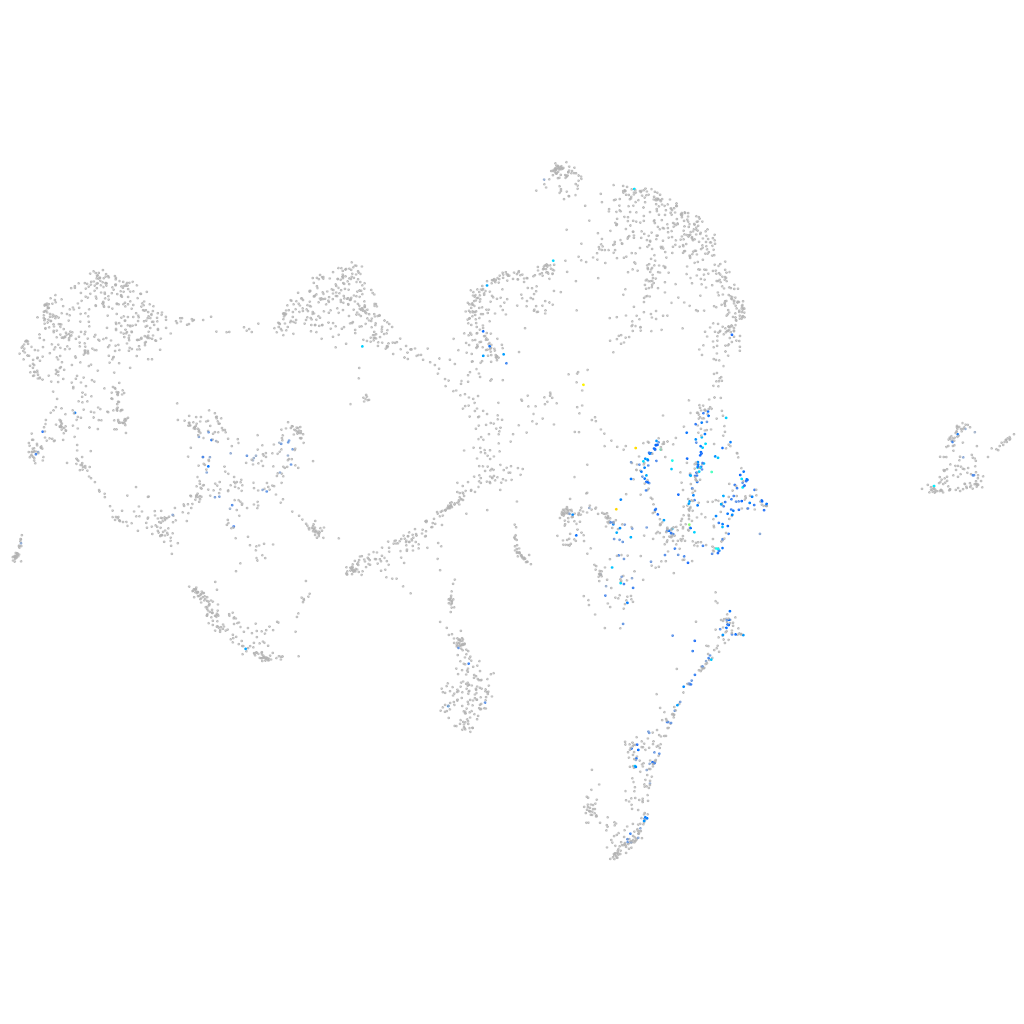
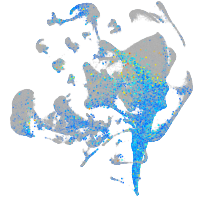
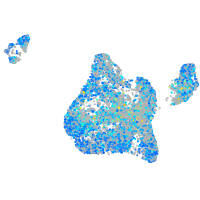
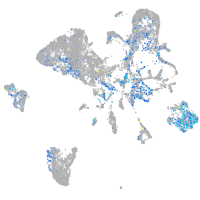
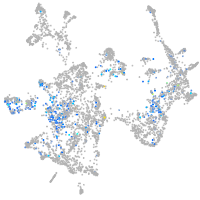
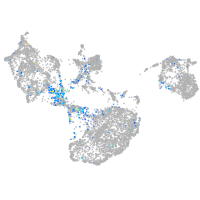
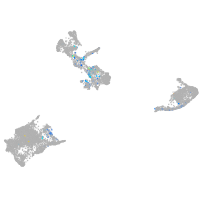
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrm2 | 0.478 | COX3 | -0.190 |
| zgc:110540 | 0.451 | BX000438.2 | -0.180 |
| cks1b | 0.411 | mt-co2 | -0.179 |
| mibp | 0.411 | tob1b | -0.174 |
| stmn1a | 0.407 | ppdpfb | -0.159 |
| cdca5 | 0.407 | selenow1 | -0.158 |
| dut | 0.405 | nfe2l2a | -0.157 |
| rrm1 | 0.405 | si:dkey-248g15.3 | -0.157 |
| slbp | 0.403 | soul2 | -0.155 |
| pcna | 0.401 | zgc:153284 | -0.155 |
| dek | 0.397 | si:dkey-247k7.2 | -0.151 |
| LOC100330864 | 0.389 | abcb5 | -0.150 |
| nasp | 0.387 | aldob | -0.148 |
| h2afx | 0.386 | gstp1 | -0.147 |
| chaf1a | 0.383 | mt-co1 | -0.146 |
| fbxo5 | 0.382 | sod1 | -0.145 |
| mis12 | 0.381 | si:ch211-195b11.3 | -0.143 |
| smc4 | 0.379 | sqstm1 | -0.142 |
| smc2 | 0.372 | selenow2b | -0.139 |
| zgc:110216 | 0.371 | tdh | -0.139 |
| ccna2 | 0.371 | mgst1.2 | -0.138 |
| rpa2 | 0.369 | zgc:110333 | -0.137 |
| tk1 | 0.363 | rplp2 | -0.136 |
| lig1 | 0.363 | itm2bb | -0.136 |
| lbr | 0.362 | icn2 | -0.136 |
| mki67 | 0.360 | pvalb1 | -0.134 |
| prim2 | 0.358 | ppl | -0.132 |
| hmgb2b | 0.356 | zgc:193505 | -0.132 |
| ncapg | 0.355 | eef1da | -0.132 |
| tubb2b | 0.353 | gsto2 | -0.131 |
| dhfr | 0.352 | nupr1a | -0.130 |
| cbx3a | 0.350 | si:dkey-193p11.2 | -0.129 |
| CABZ01005379.1 | 0.348 | fbp1a | -0.128 |
| h3f3a | 0.347 | enosf1 | -0.128 |
| rpa3 | 0.343 | epdl1 | -0.128 |