kelch repeat-containing protein
ZFIN 
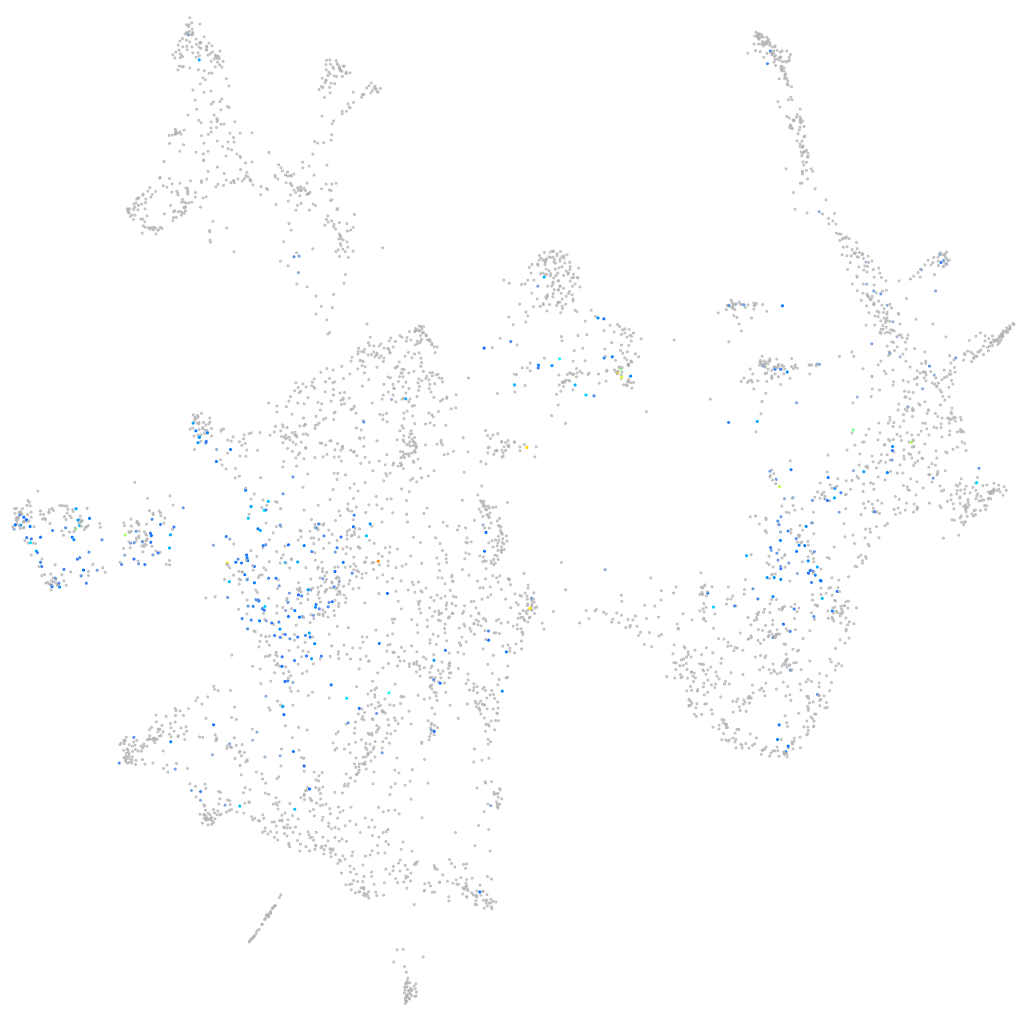
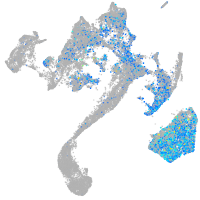
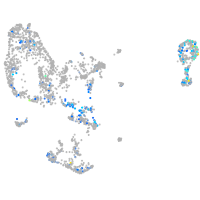
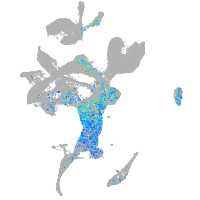
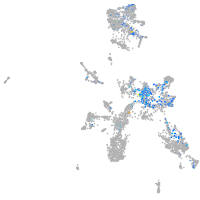
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrm2 | 0.407 | h1f0 | -0.123 |
| zgc:110540 | 0.396 | atp2b1a | -0.113 |
| mibp | 0.388 | tob1b | -0.109 |
| rrm1 | 0.381 | COX3 | -0.108 |
| pcna | 0.377 | calm1a | -0.102 |
| cdca5 | 0.350 | ip6k2a | -0.101 |
| rpa2 | 0.346 | CR383676.1 | -0.099 |
| cks1b | 0.342 | atp6v1e1b | -0.097 |
| dut | 0.340 | otofb | -0.092 |
| CABZ01005379.1 | 0.340 | b2ml | -0.092 |
| slbp | 0.339 | mt-co2 | -0.092 |
| asf1ba | 0.335 | pvalb1 | -0.091 |
| chaf1a | 0.332 | nupr1a | -0.091 |
| LOC100330864 | 0.332 | mt-co1 | -0.089 |
| stmn1a | 0.329 | tmem59 | -0.089 |
| esco2 | 0.329 | ccni | -0.089 |
| si:dkey-6i22.5 | 0.323 | calm1b | -0.089 |
| dek | 0.322 | eno1a | -0.088 |
| fbxo5 | 0.321 | hist2h2l | -0.088 |
| zgc:110216 | 0.321 | atp1a3b | -0.087 |
| dhfr | 0.320 | calml4a | -0.087 |
| lig1 | 0.310 | eef1da | -0.086 |
| prim2 | 0.306 | cd164l2 | -0.086 |
| suv39h1b | 0.303 | pkig | -0.086 |
| unga | 0.302 | col9a1a | -0.086 |
| sumo3b | 0.302 | dedd1 | -0.086 |
| rfc4 | 0.302 | kif1aa | -0.086 |
| fen1 | 0.301 | apba1a | -0.085 |
| ncapg | 0.298 | lrrc73 | -0.085 |
| ccna2 | 0.294 | gapdhs | -0.085 |
| rpa3 | 0.289 | tspan13b | -0.084 |
| dnajc9 | 0.288 | nptna | -0.084 |
| ncapd2 | 0.287 | emb | -0.084 |
| shmt1 | 0.284 | CR925719.1 | -0.084 |
| smc2 | 0.284 | rnasekb | -0.084 |