kinesin family member 6
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 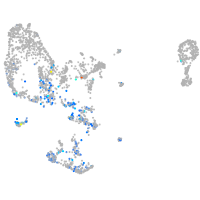 |
neural |
spinal cord / glia |
eye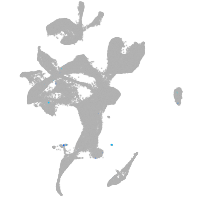 |
taste / olfactory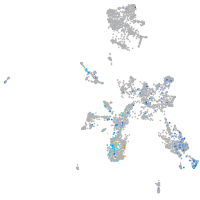 |
otic / lateral line |
epidermis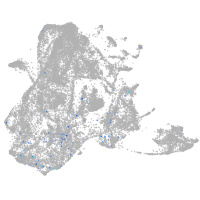 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ncs1a | 0.890 | gar1 | -0.203 |
| sept5b | 0.826 | rps18 | -0.201 |
| si:dkey-78a14.4 | 0.800 | ddx23 | -0.196 |
| si:ch73-127m5.1 | 0.726 | hp1bp3 | -0.194 |
| slc25a1b | 0.726 | si:ch211-214j24.10 | -0.194 |
| nlrc3 | 0.726 | marcksb | -0.193 |
| rgra | 0.694 | acin1b | -0.189 |
| r3hdm4 | 0.662 | ncl | -0.185 |
| XRCC2 | 0.655 | zgc:111986 | -0.179 |
| CU633991.2 | 0.651 | marcksl1b | -0.175 |
| il6r | 0.651 | ndufs5 | -0.172 |
| fgf12a | 0.638 | top1l | -0.169 |
| abo | 0.636 | rpl29 | -0.168 |
| atg16l2 | 0.636 | thoc2 | -0.167 |
| BX927130.4 | 0.636 | smarca5 | -0.166 |
| c1ql3a | 0.636 | baz1b | -0.164 |
| CABZ01066320.2 | 0.636 | thoc7 | -0.163 |
| calb1 | 0.636 | luc7l3 | -0.162 |
| CR354610.1 | 0.636 | dynll2a | -0.160 |
| CR385024.1 | 0.636 | srfbp1 | -0.158 |
| cx23 | 0.636 | npm1a | -0.154 |
| ftr57 | 0.636 | ttf1 | -0.154 |
| gcdhb | 0.636 | lig1 | -0.153 |
| GRIK2 | 0.636 | ccnb1 | -0.153 |
| itga3a | 0.636 | larp7 | -0.151 |
| itgb3b | 0.636 | srsf1a | -0.150 |
| kiss2 | 0.636 | tgif1 | -0.149 |
| LOC101883326 | 0.636 | lyar | -0.149 |
| LOC103909855 | 0.636 | si:dkeyp-69c1.6 | -0.147 |
| LOC103910921 | 0.636 | chd4b | -0.147 |
| LOC108179274 | 0.636 | zcchc17 | -0.146 |
| LOC562049 | 0.636 | atp6v0ca | -0.146 |
| map1sb | 0.636 | gnl3 | -0.144 |
| notum1b | 0.636 | srsf5a | -0.142 |
| PCP4 | 0.636 | si:dkeyp-87e7.4 | -0.141 |