kinesin family member 6
ZFIN 
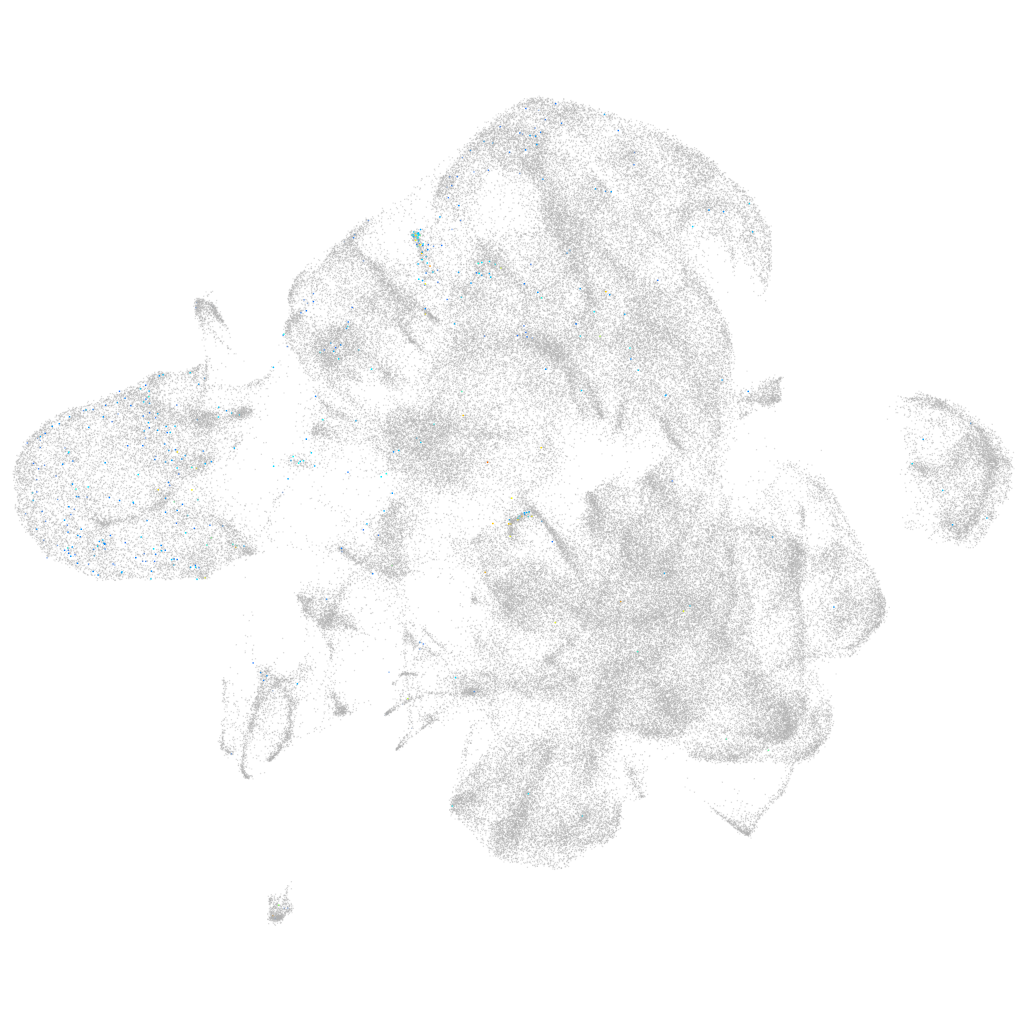
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 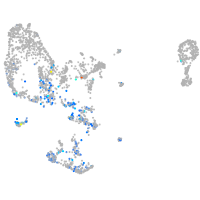 |
neural |
spinal cord / glia |
eye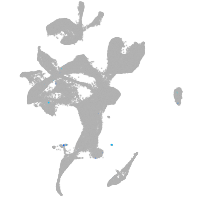 |
taste / olfactory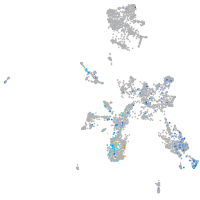 |
otic / lateral line |
epidermis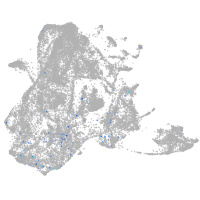 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.124 | ptmaa | -0.040 |
| ccdc173 | 0.091 | elavl3 | -0.039 |
| foxj1a | 0.085 | tuba1c | -0.037 |
| dydc2 | 0.084 | stmn1b | -0.035 |
| dnaaf1 | 0.081 | rtn1a | -0.034 |
| enkur | 0.080 | fabp3 | -0.033 |
| nme5 | 0.077 | tmsb4x | -0.032 |
| ropn1l | 0.076 | h3f3a | -0.032 |
| si:ch211-226m7.4 | 0.076 | rpl37 | -0.032 |
| spag8 | 0.076 | gpm6ab | -0.031 |
| tex9 | 0.074 | rps10 | -0.031 |
| fabp7b | 0.073 | zgc:114188 | -0.031 |
| hepacama | 0.069 | tmsb | -0.030 |
| rsph3 | 0.068 | ppiab | -0.029 |
| bicc2 | 0.067 | COX3 | -0.028 |
| mir429b | 0.067 | ywhah | -0.028 |
| si:ch211-66e2.5 | 0.066 | hnrnpa0l | -0.028 |
| pacrg | 0.065 | gpm6aa | -0.028 |
| ttc25 | 0.064 | tubb5 | -0.028 |
| cfap126 | 0.064 | elavl4 | -0.027 |
| ribc2 | 0.064 | jpt1b | -0.026 |
| si:dkey-27p23.3 | 0.064 | nova2 | -0.026 |
| zgc:114181 | 0.063 | gng3 | -0.026 |
| cfap157 | 0.063 | ckbb | -0.026 |
| rspo3 | 0.063 | sncb | -0.025 |
| LOC103910489 | 0.063 | rtn1b | -0.025 |
| bmp6 | 0.062 | gng2 | -0.024 |
| tbata | 0.062 | mt-nd1 | -0.024 |
| drc1 | 0.062 | stmn2a | -0.024 |
| rab36 | 0.062 | myt1b | -0.024 |
| si:dkey-42p14.3 | 0.062 | si:dkey-276j7.1 | -0.024 |
| smkr1 | 0.061 | marcksl1a | -0.024 |
| efcab1 | 0.061 | atp6v0cb | -0.024 |
| rspo1 | 0.060 | celf2 | -0.023 |
| igfbp7 | 0.060 | CR383676.1 | -0.023 |