kinesin family member 26Ab
ZFIN 
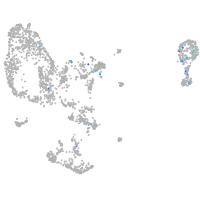
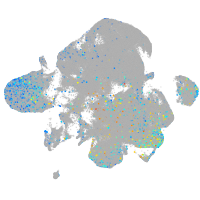
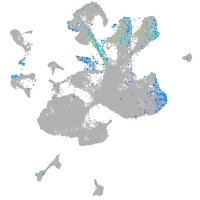
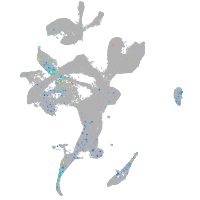
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hes6 | 0.259 | fabp3 | -0.216 |
| cx43.4 | 0.254 | actc1b | -0.184 |
| cdx4 | 0.252 | eef1da | -0.169 |
| itm2cb | 0.240 | vdac3 | -0.168 |
| nid2a | 0.237 | tpi1b | -0.166 |
| cyp26a1 | 0.235 | bhmt | -0.160 |
| vox | 0.233 | col1a2 | -0.156 |
| fgf8a | 0.224 | pvalb1 | -0.155 |
| hspb1 | 0.218 | ckma | -0.154 |
| nradd | 0.217 | pvalb2 | -0.153 |
| cirbpa | 0.214 | si:dkey-16p21.8 | -0.152 |
| hmga1a | 0.214 | acta1b | -0.152 |
| greb1 | 0.212 | ckmb | -0.151 |
| apoeb | 0.211 | tpma | -0.150 |
| nucks1a | 0.210 | gamt | -0.150 |
| ncl | 0.210 | eno1a | -0.150 |
| hnrnpaba | 0.209 | aldoab | -0.149 |
| BX927258.1 | 0.209 | col1a1a | -0.147 |
| gdf11 | 0.209 | ak1 | -0.147 |
| khdrbs1a | 0.209 | sparc | -0.147 |
| nop58 | 0.208 | atp2a1 | -0.146 |
| syncrip | 0.207 | eif4a1b | -0.140 |
| hnrnpabb | 0.206 | eno3 | -0.139 |
| her7 | 0.206 | tnnc2 | -0.138 |
| nop56 | 0.205 | CABZ01078594.1 | -0.138 |
| sp5l | 0.204 | gapdh | -0.138 |
| setb | 0.204 | actn3a | -0.138 |
| hmgn7 | 0.204 | tnni2a.4 | -0.138 |
| seta | 0.203 | tmem38a | -0.137 |
| hoxb10a | 0.203 | nme2b.2 | -0.136 |
| fgf3 | 0.202 | col1a1b | -0.136 |
| ptges3b | 0.202 | atp5if1b | -0.135 |
| si:ch73-281n10.2 | 0.201 | smyd1a | -0.135 |
| h2afvb | 0.201 | mdh2 | -0.135 |
| npm1a | 0.200 | pgk1 | -0.134 |