kinesin family member 22
ZFIN 
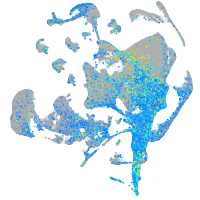
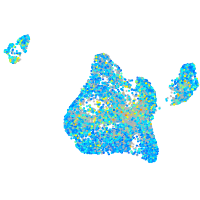
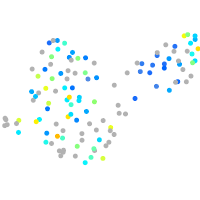
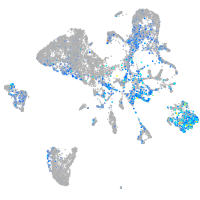
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.551 | gamt | -0.293 |
| tpx2 | 0.519 | gatm | -0.265 |
| mad2l1 | 0.504 | rpl37 | -0.261 |
| ccnb1 | 0.494 | gapdh | -0.259 |
| cdca8 | 0.490 | zgc:92744 | -0.251 |
| ube2c | 0.483 | bhmt | -0.249 |
| plk1 | 0.482 | rps10 | -0.242 |
| ccna2 | 0.482 | ahcy | -0.238 |
| hmgb2a | 0.479 | gpx4a | -0.234 |
| aurkb | 0.461 | zgc:114188 | -0.231 |
| nusap1 | 0.460 | pnp4b | -0.229 |
| cdk1 | 0.459 | apoa1b | -0.227 |
| seta | 0.457 | nupr1b | -0.221 |
| anp32b | 0.456 | fbp1b | -0.219 |
| hmga1a | 0.446 | apoa4b.1 | -0.218 |
| dlgap5 | 0.445 | apoa2 | -0.217 |
| hmgb2b | 0.443 | mat1a | -0.215 |
| cbx3a | 0.439 | agxtb | -0.213 |
| anp32a | 0.437 | eno3 | -0.213 |
| cenpf | 0.436 | pklr | -0.212 |
| dek | 0.435 | afp4 | -0.211 |
| top2a | 0.434 | rps17 | -0.204 |
| si:ch73-281n10.2 | 0.434 | sod1 | -0.202 |
| si:ch211-69g19.2 | 0.432 | rbp4 | -0.201 |
| prc1a | 0.430 | suclg1 | -0.200 |
| kmt5ab | 0.429 | abat | -0.199 |
| kifc1 | 0.428 | grhprb | -0.199 |
| hnrnpa1b | 0.423 | scp2a | -0.198 |
| hnrnpabb | 0.421 | aqp12 | -0.198 |
| aurka | 0.421 | g6pca.2 | -0.197 |
| si:ch211-288g17.3 | 0.421 | apoc2 | -0.196 |
| hnrnpa0b | 0.421 | glud1b | -0.196 |
| cks1b | 0.421 | dap | -0.196 |
| tubb2b | 0.421 | fetub | -0.196 |
| syncrip | 0.419 | cx32.3 | -0.194 |