kinesin family member 22
ZFIN 
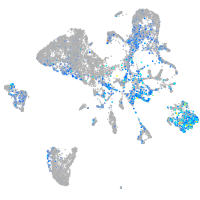
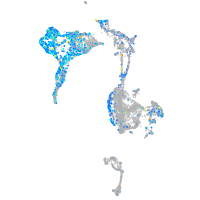
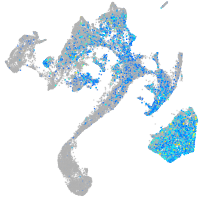
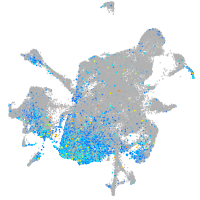
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccna2 | 0.531 | txn | -0.227 |
| aurkb | 0.524 | gstp1 | -0.216 |
| tpx2 | 0.521 | anxa5b | -0.210 |
| cdk1 | 0.518 | gapdhs | -0.207 |
| mad2l1 | 0.518 | wu:fb18f06 | -0.192 |
| mki67 | 0.516 | selenow2b | -0.191 |
| cks1b | 0.505 | tmem59 | -0.191 |
| cdc20 | 0.502 | cldnh | -0.188 |
| ube2c | 0.500 | calm1a | -0.185 |
| si:ch211-69g19.2 | 0.497 | fxyd1 | -0.177 |
| nusap1 | 0.497 | nfe2l2a | -0.176 |
| top2a | 0.487 | itm2ba | -0.173 |
| kif11 | 0.482 | selenow1 | -0.169 |
| cdca8 | 0.481 | rnasekb | -0.169 |
| spdl1 | 0.476 | chga | -0.169 |
| aspm | 0.474 | vamp2 | -0.167 |
| cenpf | 0.469 | bik | -0.165 |
| ccnb1 | 0.465 | syt1a | -0.160 |
| kifc1 | 0.462 | CR383676.1 | -0.160 |
| birc5a | 0.459 | gabarapa | -0.156 |
| prc1a | 0.458 | COX3 | -0.155 |
| lbr | 0.458 | scg2a | -0.154 |
| kpna2 | 0.456 | pvalb5 | -0.154 |
| ncapg | 0.455 | ponzr6 | -0.152 |
| cks2 | 0.455 | camk2n1a | -0.152 |
| kif23 | 0.451 | si:ch211-153b23.5 | -0.151 |
| aurka | 0.450 | scg2b | -0.151 |
| smc2 | 0.442 | rtn1a | -0.151 |
| kmt5ab | 0.435 | calr | -0.151 |
| armc1l | 0.435 | mt-co2 | -0.148 |
| plk1 | 0.434 | calb2a | -0.148 |
| cdca5 | 0.434 | tpi1b | -0.147 |
| numa1 | 0.432 | msi2b | -0.146 |
| tacc3 | 0.431 | atp6v1e1b | -0.146 |
| g2e3 | 0.426 | b2ml | -0.144 |