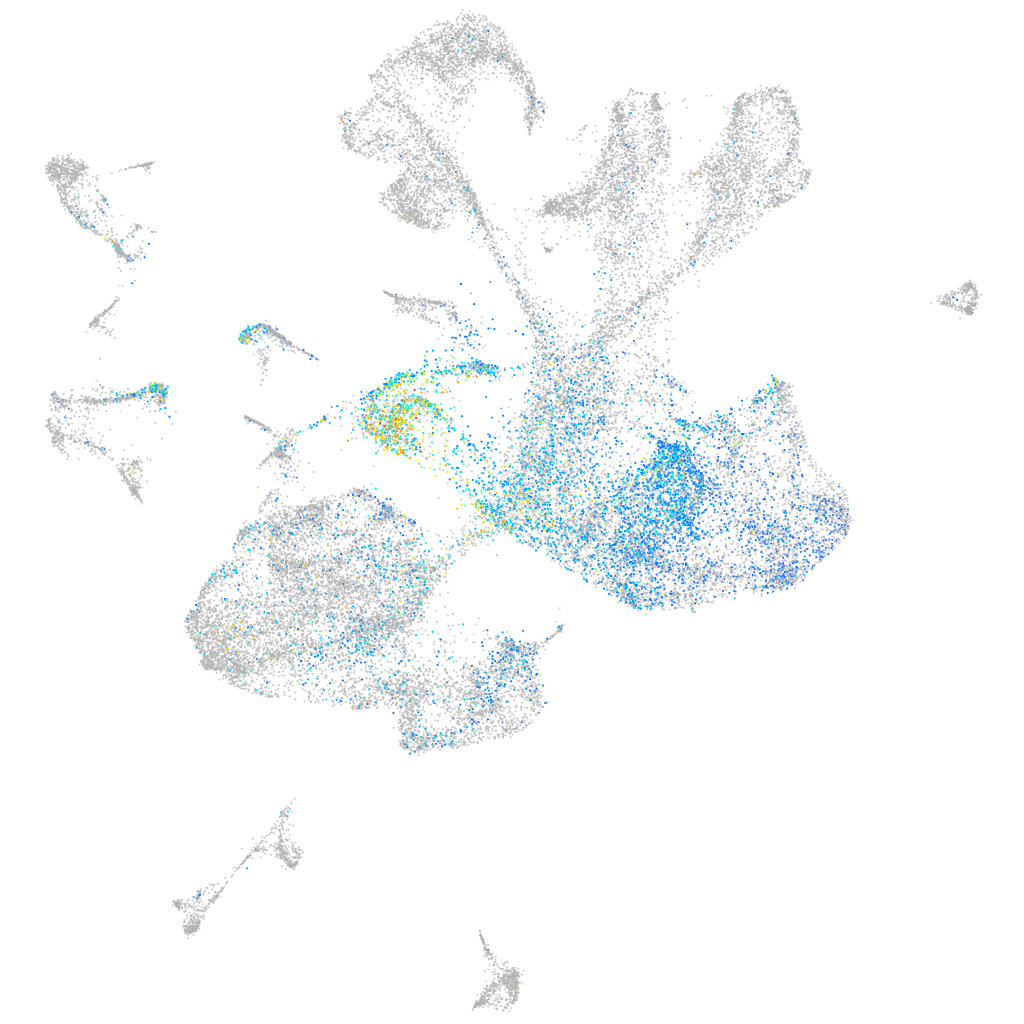
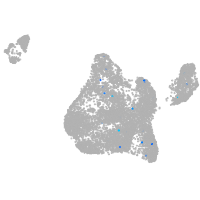
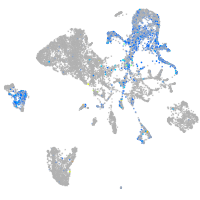
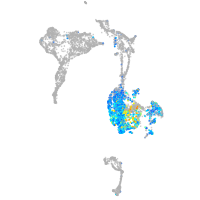
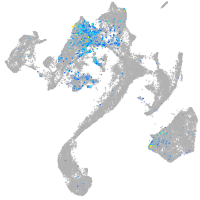
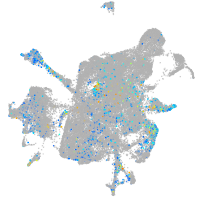
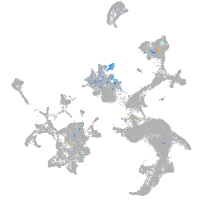
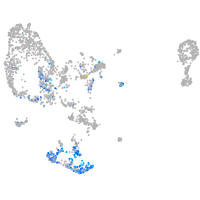
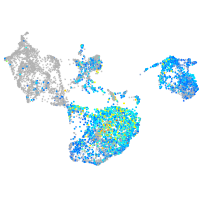
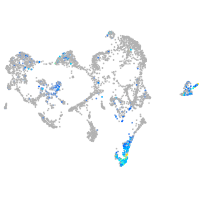
"integrin, beta 4"
ZFIN 
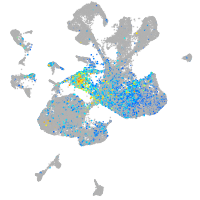
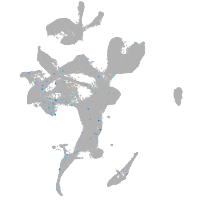
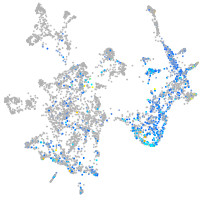
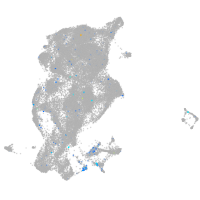
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gfap | 0.438 | stmn1b | -0.174 |
| col4a5 | 0.381 | gpm6ab | -0.168 |
| col4a6 | 0.376 | rtn1a | -0.162 |
| cldn7a | 0.337 | gng3 | -0.160 |
| cav1 | 0.323 | ywhah | -0.159 |
| plecb | 0.313 | elavl3 | -0.158 |
| wu:fi04e12 | 0.300 | stxbp1a | -0.156 |
| pleca | 0.295 | elavl4 | -0.154 |
| clstn2 | 0.289 | vamp2 | -0.154 |
| si:ch73-265d7.2 | 0.281 | sncb | -0.151 |
| vwa7 | 0.278 | atp6v0cb | -0.150 |
| zgc:113531 | 0.277 | snap25a | -0.146 |
| endouc | 0.275 | stx1b | -0.145 |
| pbxip1b | 0.275 | rnasekb | -0.143 |
| igfbp7 | 0.268 | rtn1b | -0.140 |
| vim | 0.266 | mllt11 | -0.140 |
| slc6a9 | 0.254 | tmem59l | -0.139 |
| zgc:112437 | 0.248 | syt2a | -0.135 |
| si:ch211-251b21.1 | 0.247 | nsg2 | -0.135 |
| slc10a1 | 0.244 | ywhag2 | -0.134 |
| si:dkey-245n4.2 | 0.244 | myt1b | -0.133 |
| tes | 0.239 | cplx2 | -0.133 |
| cav2 | 0.239 | calm1a | -0.132 |
| pkp3b | 0.235 | stmn2a | -0.132 |
| tgm1l1 | 0.234 | zgc:65894 | -0.132 |
| lrrc39 | 0.233 | csdc2a | -0.130 |
| cavin1b | 0.231 | map1aa | -0.129 |
| rgs5a | 0.226 | gap43 | -0.128 |
| mdka | 0.220 | ptmaa | -0.128 |
| slc4a10a | 0.218 | gng2 | -0.128 |
| dsg2.1 | 0.217 | atpv0e2 | -0.127 |
| pkd1b | 0.216 | jagn1a | -0.126 |
| col18a1a | 0.212 | rbfox1 | -0.126 |
| adamts3 | 0.208 | aplp1 | -0.125 |
| crip2 | 0.208 | onecut1 | -0.124 |