inka box actin regulator 1a
ZFIN 
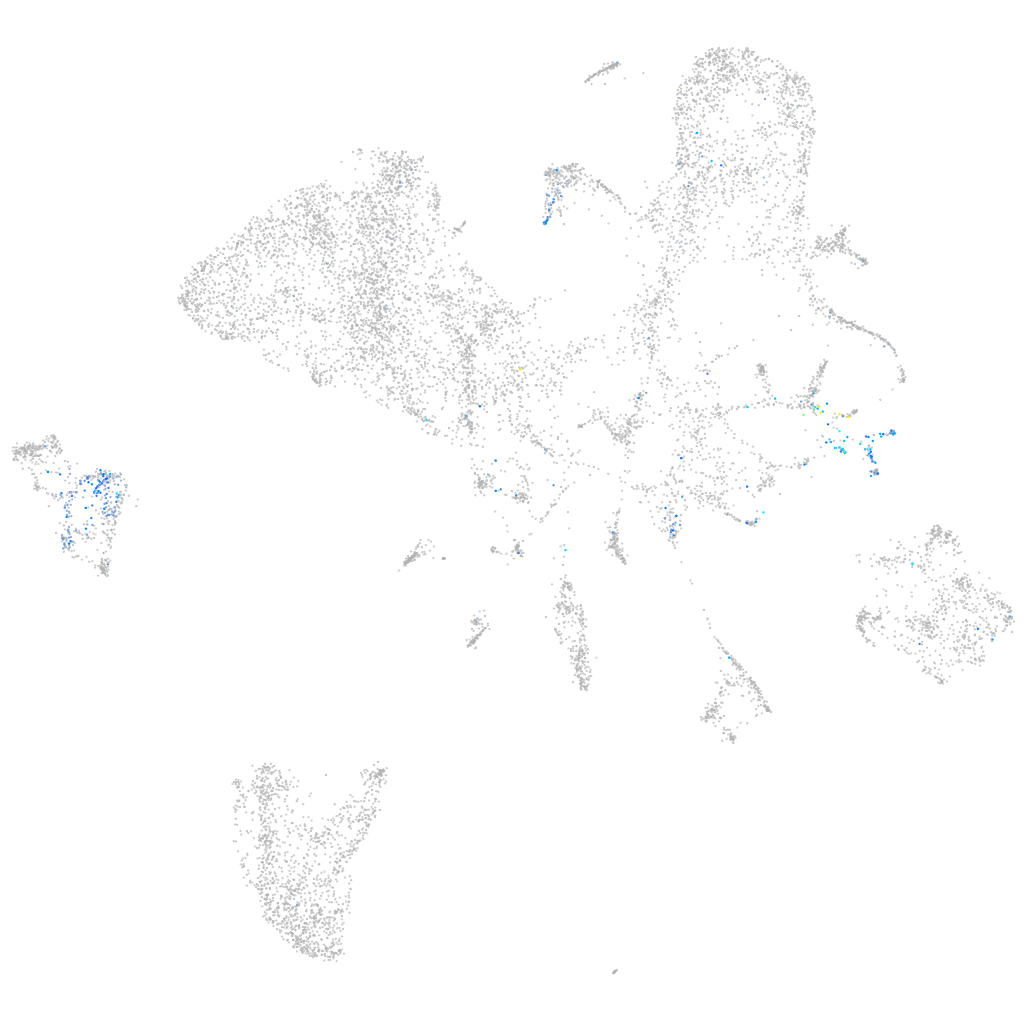
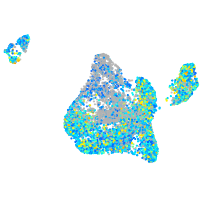
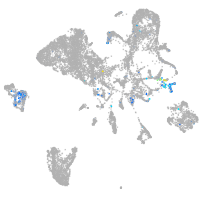
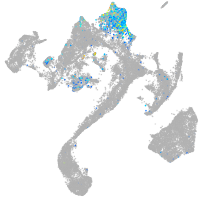
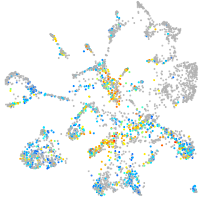
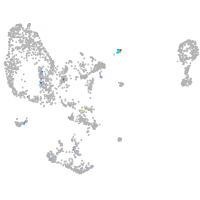
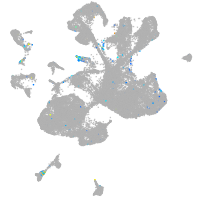
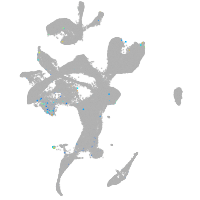
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-153k10.9 | 0.315 | gapdh | -0.166 |
| isl1 | 0.312 | gamt | -0.145 |
| ndufa4l2a | 0.307 | gatm | -0.133 |
| insm1a | 0.303 | fbp1b | -0.128 |
| scg2a | 0.302 | bhmt | -0.123 |
| scgn | 0.300 | aldob | -0.123 |
| neurod1 | 0.280 | gpx4a | -0.120 |
| kcnj11 | 0.275 | glud1b | -0.120 |
| SLC5A10 | 0.274 | mat1a | -0.117 |
| abhd15a | 0.274 | apoa4b.1 | -0.115 |
| cldni | 0.268 | apoa1b | -0.113 |
| mir7a-1 | 0.261 | apoc2 | -0.113 |
| ins | 0.258 | ahcy | -0.113 |
| dkk3b | 0.258 | cx32.3 | -0.110 |
| scg3 | 0.255 | afp4 | -0.110 |
| nkx2.2a | 0.250 | scp2a | -0.108 |
| pax6b | 0.249 | gstt1a | -0.105 |
| gapdhs | 0.237 | eno3 | -0.104 |
| gpr22a | 0.234 | agxta | -0.104 |
| btc | 0.233 | gcshb | -0.103 |
| gip | 0.232 | aqp12 | -0.102 |
| hapln1a | 0.232 | nupr1b | -0.102 |
| tp63 | 0.230 | agxtb | -0.101 |
| fam49bb | 0.229 | sult2st2 | -0.101 |
| pcsk1nl | 0.227 | g6pca.2 | -0.100 |
| sst2 | 0.225 | apoc1 | -0.100 |
| LOC100537384 | 0.225 | pklr | -0.099 |
| zgc:153911 | 0.217 | aldh7a1 | -0.099 |
| hdac9b | 0.217 | gstr | -0.098 |
| syt1a | 0.215 | suclg1 | -0.098 |
| rprmb | 0.212 | grhprb | -0.098 |
| tspan7b | 0.212 | cox7a1 | -0.098 |
| LOC571123 | 0.211 | suclg2 | -0.097 |
| slc35g2b | 0.211 | si:dkey-16p21.8 | -0.097 |
| itga6b | 0.208 | tfa | -0.097 |