inner mitochondrial membrane peptidase subunit 2
ZFIN 
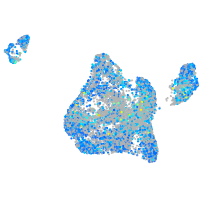
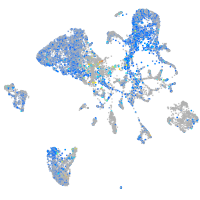
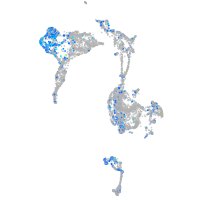
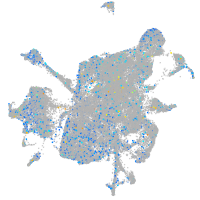
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ldhba | 0.226 | si:ch73-1a9.3 | -0.189 |
| atp5meb | 0.211 | marcksb | -0.189 |
| si:dkey-33c12.12 | 0.206 | ptmab | -0.186 |
| atp5mc3b | 0.205 | apoeb | -0.182 |
| si:dkey-16p21.8 | 0.204 | marcksl1b | -0.178 |
| uqcrc1 | 0.202 | hspb1 | -0.173 |
| dlst | 0.199 | wu:fb97g03 | -0.172 |
| pdha1a | 0.197 | apoc1 | -0.169 |
| atp5fa1 | 0.197 | nucks1a | -0.158 |
| idh2 | 0.193 | si:ch73-281n10.2 | -0.157 |
| atp1b1a | 0.192 | anp32a | -0.157 |
| vdac3 | 0.191 | hnrnpabb | -0.155 |
| cox4i1 | 0.191 | NC-002333.4 | -0.152 |
| cox6a1 | 0.190 | hmgb2a | -0.151 |
| pcbd1 | 0.189 | ilf3b | -0.150 |
| rps10 | 0.189 | hnrnpaba | -0.149 |
| ddt | 0.189 | hmgn6 | -0.149 |
| sod2 | 0.187 | cdx4 | -0.149 |
| slc25a5 | 0.187 | hmga1a | -0.148 |
| COX5B | 0.187 | acin1a | -0.146 |
| zgc:162944 | 0.187 | syncrip | -0.144 |
| gpx1a | 0.184 | akap12b | -0.144 |
| suclg1 | 0.183 | pabpc1a | -0.143 |
| eno3 | 0.183 | msna | -0.142 |
| mdh2 | 0.183 | si:ch211-288g17.3 | -0.141 |
| GCA | 0.183 | si:ch211-222l21.1 | -0.141 |
| cdh17 | 0.182 | top1l | -0.140 |
| ndufb10 | 0.182 | ncl | -0.137 |
| zgc:162730 | 0.181 | h3f3d | -0.137 |
| as3mt | 0.181 | snu13b | -0.136 |
| cox8a | 0.181 | fbl | -0.134 |
| mgst3a | 0.180 | ppig | -0.134 |
| cdaa | 0.179 | hes6 | -0.134 |
| vill | 0.176 | anp32b | -0.132 |
| atp5mc1 | 0.176 | hnrnpa1a | -0.132 |