immunoglobulin mu DNA binding protein 2
ZFIN 
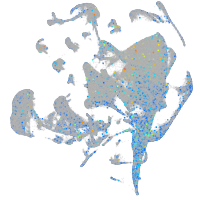
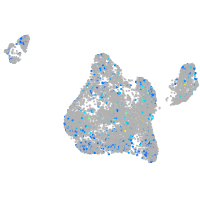
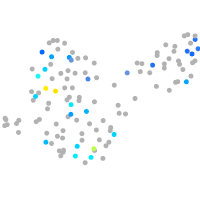
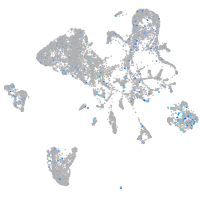
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.065 | ptmaa | -0.050 |
| nop58 | 0.065 | rpl37 | -0.040 |
| npm1a | 0.063 | tmsb4x | -0.040 |
| nop2 | 0.062 | rtn1a | -0.038 |
| si:ch211-152c2.3 | 0.062 | rps10 | -0.037 |
| nop56 | 0.061 | tuba1c | -0.037 |
| ncl | 0.059 | gpm6aa | -0.036 |
| dkc1 | 0.059 | stmn1b | -0.035 |
| fbl | 0.059 | gpm6ab | -0.034 |
| zgc:110425 | 0.058 | CR383676.1 | -0.034 |
| apoeb | 0.058 | elavl3 | -0.033 |
| bms1 | 0.056 | sncb | -0.033 |
| mki67 | 0.056 | zgc:114188 | -0.033 |
| akap12b | 0.056 | marcksl1a | -0.031 |
| nr6a1a | 0.056 | ckbb | -0.031 |
| pprc1 | 0.055 | atp6v0cb | -0.031 |
| anp32b | 0.055 | atpv0e2 | -0.031 |
| gnl3 | 0.055 | rnasekb | -0.031 |
| hnrnpa1b | 0.055 | pvalb1 | -0.030 |
| rsl1d1 | 0.055 | calm1a | -0.030 |
| lig1 | 0.054 | cspg5a | -0.030 |
| gar1 | 0.054 | elavl4 | -0.030 |
| stm | 0.053 | rtn1b | -0.030 |
| ebna1bp2 | 0.053 | zgc:158463 | -0.029 |
| hmga1a | 0.053 | h3f3a | -0.029 |
| eif5a2 | 0.053 | ywhag2 | -0.029 |
| pes | 0.053 | pvalb2 | -0.029 |
| NC-002333.4 | 0.052 | COX3 | -0.028 |
| tpx2 | 0.052 | hbbe1.3 | -0.028 |
| ppig | 0.052 | vamp2 | -0.028 |
| eif4bb | 0.052 | kdm6bb | -0.028 |
| mybbp1a | 0.052 | actc1b | -0.028 |
| hmgb2a | 0.051 | si:dkeyp-75h12.5 | -0.028 |
| anp32a | 0.051 | celf2 | -0.028 |
| zmp:0000000624 | 0.051 | ppiab | -0.027 |