inhibitor of DNA binding 4
ZFIN 
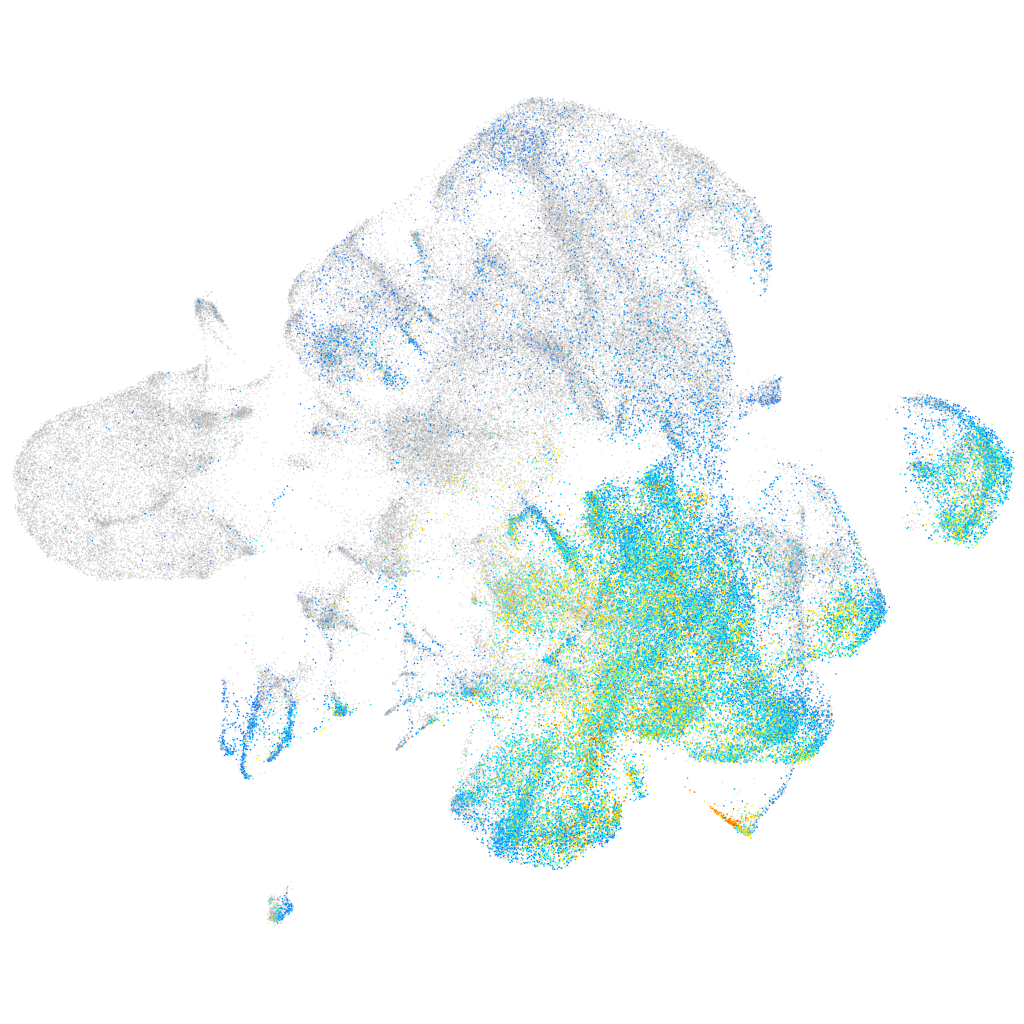
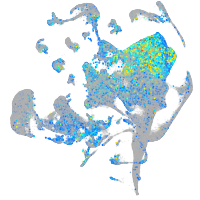
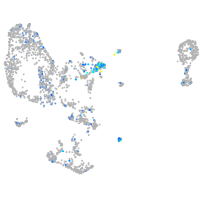
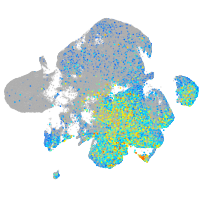
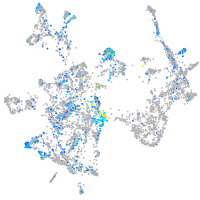
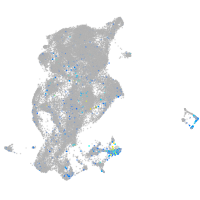
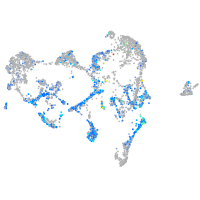
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.545 | hmgb2a | -0.480 |
| sncb | 0.519 | id1 | -0.375 |
| stmn1b | 0.499 | pcna | -0.371 |
| rtn1b | 0.486 | hmga1a | -0.359 |
| ywhag2 | 0.484 | stmn1a | -0.351 |
| stmn2a | 0.477 | hmgb2b | -0.349 |
| ckbb | 0.477 | msna | -0.338 |
| tuba1c | 0.475 | anp32b | -0.327 |
| gpm6ab | 0.474 | mdka | -0.327 |
| atp6v1e1b | 0.465 | ranbp1 | -0.322 |
| zgc:65894 | 0.463 | XLOC-003690 | -0.320 |
| atp6v0cb | 0.461 | chaf1a | -0.317 |
| gap43 | 0.458 | selenoh | -0.316 |
| atp6v1g1 | 0.456 | npm1a | -0.314 |
| mllt11 | 0.454 | si:dkey-151g10.6 | -0.314 |
| rtn1a | 0.452 | banf1 | -0.309 |
| tmsb2 | 0.450 | ccnd1 | -0.309 |
| snap25a | 0.447 | sox3 | -0.308 |
| vamp2 | 0.447 | nop58 | -0.307 |
| calm1a | 0.442 | rrm1 | -0.306 |
| zc4h2 | 0.441 | si:ch73-281n10.2 | -0.306 |
| elavl4 | 0.441 | tuba8l | -0.306 |
| stxbp1a | 0.439 | sox19a | -0.305 |
| ywhah | 0.436 | mki67 | -0.304 |
| stx1b | 0.431 | snu13b | -0.300 |
| ptmaa | 0.427 | mcm7 | -0.299 |
| calm2a | 0.424 | her15.1 | -0.297 |
| fez1 | 0.421 | tuba8l4 | -0.297 |
| rnasekb | 0.420 | XLOC-003689 | -0.297 |
| gapdhs | 0.414 | dut | -0.294 |
| si:dkeyp-75h12.5 | 0.411 | nop56 | -0.294 |
| csdc2a | 0.407 | lig1 | -0.290 |
| gng2 | 0.402 | nutf2l | -0.289 |
| si:dkey-276j7.1 | 0.399 | dkc1 | -0.289 |
| h2afx1 | 0.397 | mcm6 | -0.287 |