"heat shock protein, alpha-crystallin-related, 1"
ZFIN 
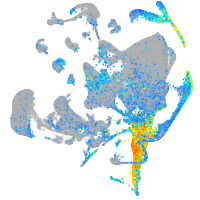
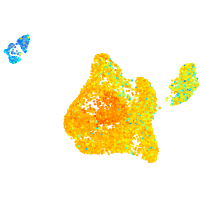
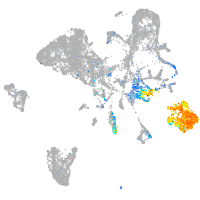
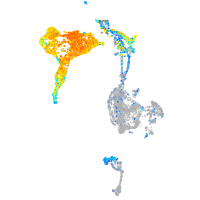
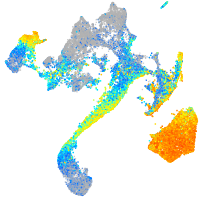
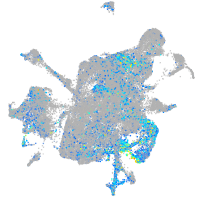
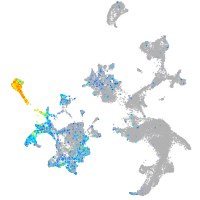
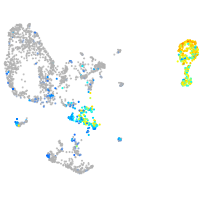
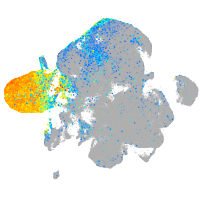
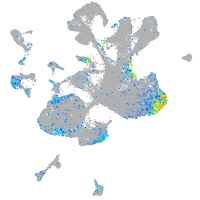
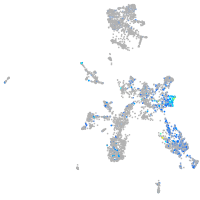
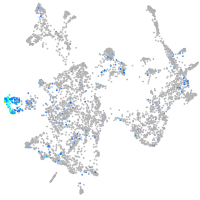
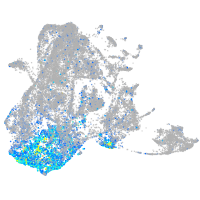
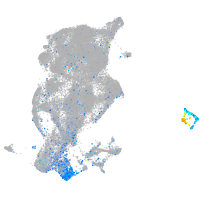
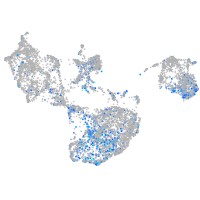
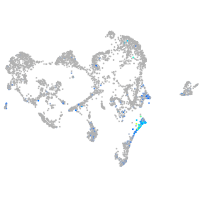
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmga1a | 0.663 | pvalb2 | -0.474 |
| hnrnpabb | 0.644 | pvalb1 | -0.474 |
| cx43.4 | 0.631 | mylpfa | -0.471 |
| hnrnpa0b | 0.607 | hbbe1.3 | -0.468 |
| hmgb2a | 0.592 | gapdhs | -0.463 |
| hnrnpaba | 0.591 | hbae1.1 | -0.448 |
| seta | 0.572 | fabp7a | -0.437 |
| snrpb | 0.559 | smdt1a | -0.419 |
| khdrbs1a | 0.547 | actc1b | -0.404 |
| hnrnpub | 0.531 | psmb8a | -0.401 |
| srsf3b | 0.520 | hbae3 | -0.398 |
| hnrnph1l | 0.515 | icn | -0.393 |
| anp32b | 0.509 | apoa2 | -0.390 |
| ctsla | 0.497 | anxa5b | -0.388 |
| apoeb | 0.496 | NDUFB1 | -0.385 |
| ccna2 | 0.491 | alcama | -0.382 |
| mphosph10 | 0.484 | cyt1 | -0.377 |
| ranbp1 | 0.470 | apoa1b | -0.376 |
| hmgb2b | 0.469 | ost4 | -0.375 |
| paip2b | 0.469 | si:ch211-132p1.3 | -0.374 |
| setb | 0.468 | stau2 | -0.373 |
| syncrip | 0.465 | ptmaa | -0.371 |
| ilf2 | 0.463 | cox5b2 | -0.368 |
| akap12b | 0.458 | syt1a | -0.368 |
| sephs1 | 0.458 | isg15 | -0.367 |
| sf3b4 | 0.454 | tdrd1 | -0.364 |
| hnrnpl2 | 0.452 | rnasekb | -0.363 |
| anp32e | 0.450 | zgc:162509 | -0.362 |
| tubb2b | 0.449 | prdx4 | -0.359 |
| khsrp | 0.446 | si:ch211-195b11.3 | -0.358 |
| ptges3b | 0.445 | krtt1c19e | -0.358 |
| mki67 | 0.442 | agrn | -0.354 |
| anp32a | 0.441 | rpl37 | -0.354 |
| stm | 0.441 | rbp4 | -0.351 |
| ddx39aa | 0.439 | serpinb1 | -0.350 |