"heat shock protein, alpha-crystallin-related, 1"
ZFIN 
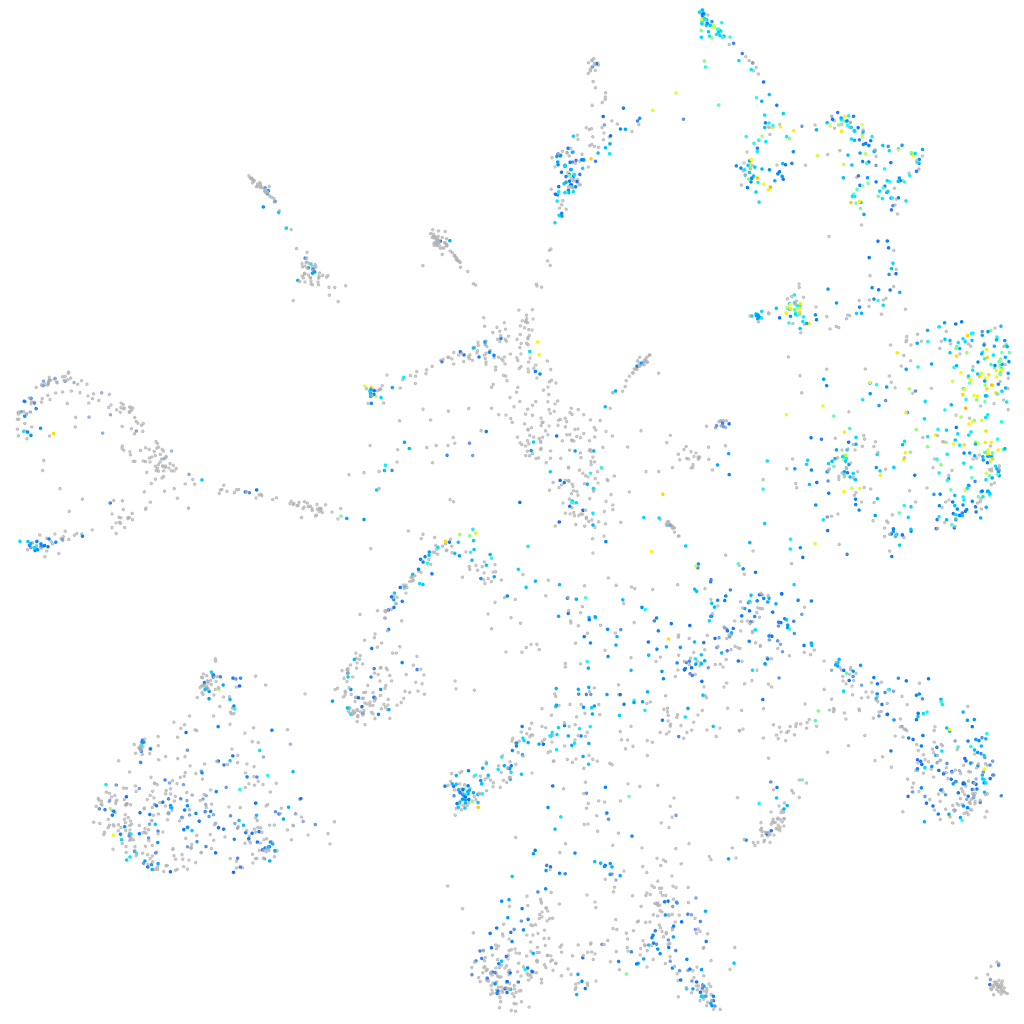
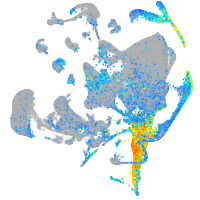
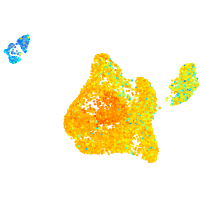
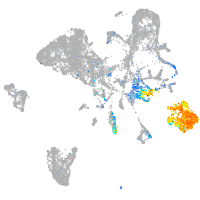
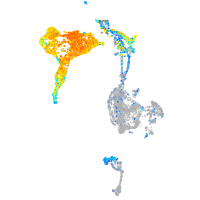
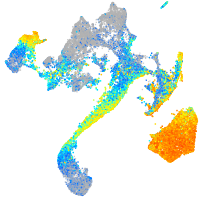
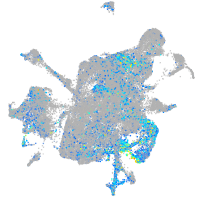
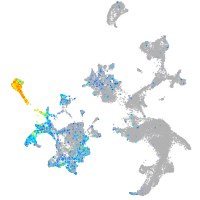
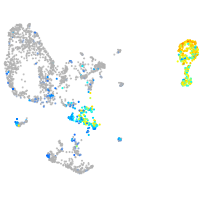
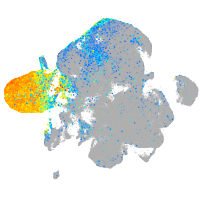
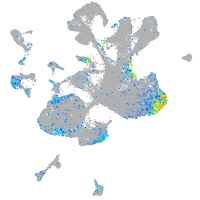
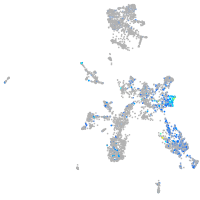
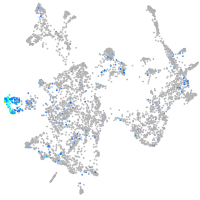
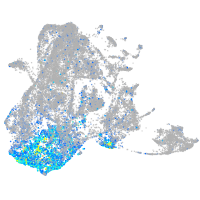
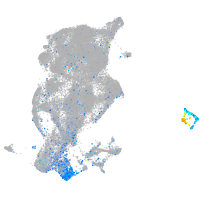
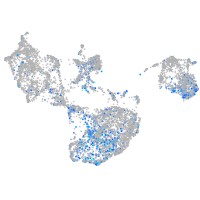
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt94 | 0.491 | tpm1 | -0.253 |
| podxl | 0.484 | acta2 | -0.248 |
| jam2b | 0.472 | myh11a | -0.243 |
| aqp1a.1 | 0.449 | BX088707.3 | -0.230 |
| tmem88b | 0.421 | tagln | -0.226 |
| cavin2b | 0.373 | mylkb | -0.217 |
| krt8 | 0.370 | lmod1b | -0.206 |
| cd151 | 0.366 | myl9a | -0.190 |
| COLEC10 | 0.363 | gapdhs | -0.186 |
| cavin1b | 0.360 | ptmaa | -0.185 |
| bcam | 0.356 | csrp1b | -0.180 |
| cav1 | 0.354 | tpm2 | -0.168 |
| rhag | 0.348 | cald1b | -0.166 |
| krt15 | 0.346 | myocd | -0.163 |
| anxa1a | 0.336 | mylka | -0.158 |
| si:ch211-156j16.1 | 0.331 | fhl2a | -0.157 |
| tgm2b | 0.329 | ldha | -0.149 |
| tmem98 | 0.322 | aldocb | -0.145 |
| ehd1b | 0.321 | cnn1b | -0.138 |
| colec11 | 0.307 | tpi1b | -0.137 |
| gstm.3 | 0.306 | si:ch211-270g19.5 | -0.134 |
| serpine3 | 0.297 | pgk1 | -0.132 |
| ftr82 | 0.295 | ak1 | -0.131 |
| lrrc15 | 0.288 | rbm24a | -0.131 |
| arl4aa | 0.288 | igfbp7 | -0.129 |
| cxadr | 0.282 | angptl5 | -0.129 |
| tjp2b | 0.281 | tmem119b | -0.128 |
| aqp8a.1 | 0.281 | pcdh18a | -0.128 |
| lurap1 | 0.281 | desmb | -0.127 |
| ecm1b | 0.279 | gpm6aa | -0.126 |
| nt5e | 0.277 | gng2 | -0.126 |
| s100a10b | 0.273 | myl6 | -0.126 |
| bnc2 | 0.269 | barx1 | -0.125 |
| phactr4a | 0.263 | BX323087.1 | -0.124 |
| ehd3 | 0.263 | fbp2 | -0.123 |