heat shock protein 12B
ZFIN 
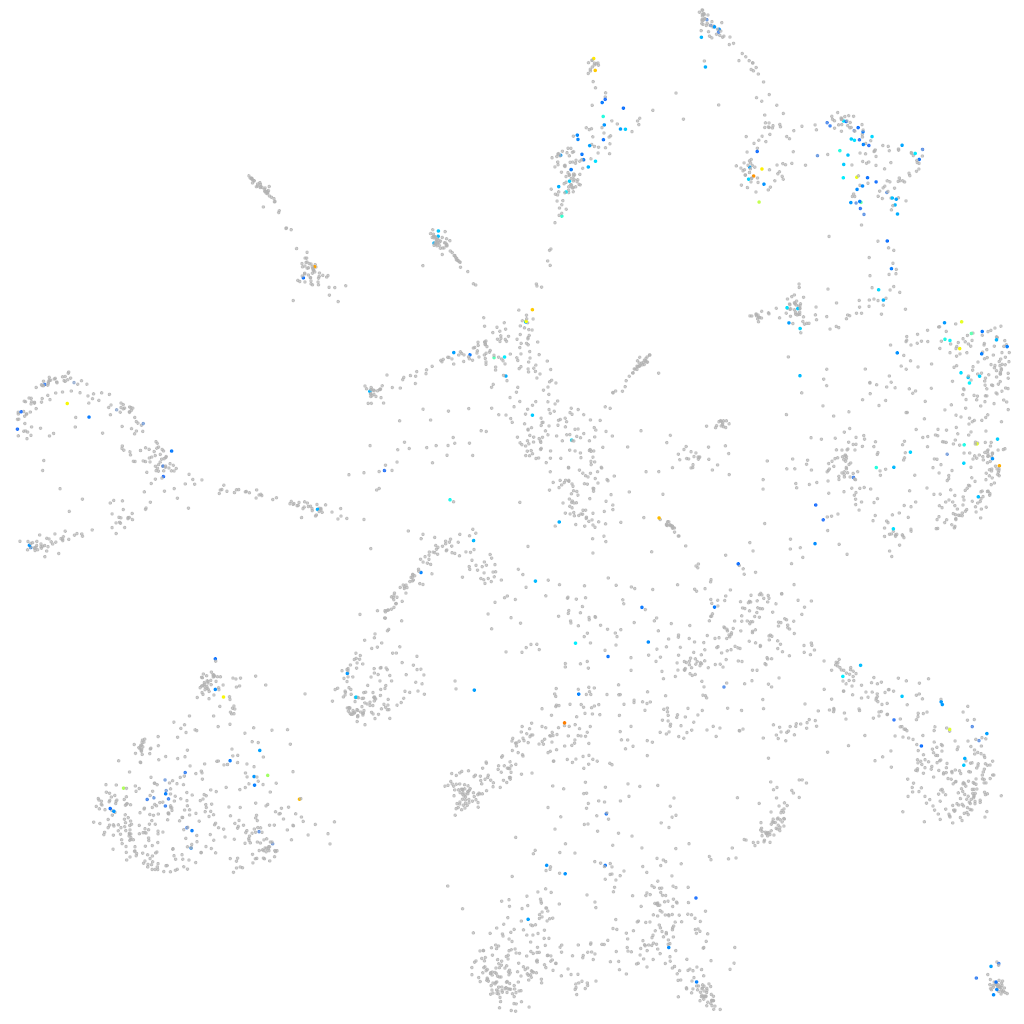
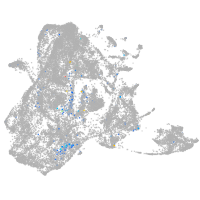
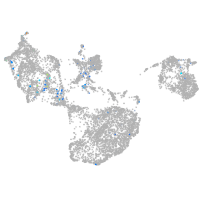
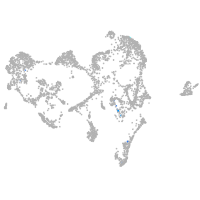
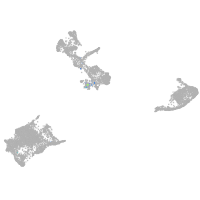
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt94 | 0.169 | BX088707.3 | -0.101 |
| si:ch211-209f23.3 | 0.165 | foxf1 | -0.093 |
| cd151 | 0.164 | mylkb | -0.083 |
| ftr82 | 0.163 | nkx2.3 | -0.081 |
| CU693479.1 | 0.163 | rbpms2a | -0.080 |
| FITM1 (1 of many) | 0.157 | tpm1 | -0.078 |
| lbx2 | 0.155 | XLOC-041870 | -0.078 |
| si:ch211-191o15.6 | 0.153 | XLOC-025423 | -0.073 |
| tnni1b | 0.149 | desmb | -0.073 |
| tbx5a | 0.147 | tpm2 | -0.072 |
| krt8 | 0.146 | BX322618.1 | -0.070 |
| anxa1a | 0.146 | BX323087.1 | -0.069 |
| tbx1 | 0.144 | ckbb | -0.068 |
| crb2a | 0.144 | inka1a | -0.067 |
| XLOC-022666 | 0.140 | myh11a | -0.066 |
| XLOC-028865 | 0.140 | si:ch211-62a1.3 | -0.066 |
| lctla | 0.139 | XLOC-028370 | -0.065 |
| slit3 | 0.138 | ntn5 | -0.064 |
| podxl | 0.138 | ptch2 | -0.064 |
| mmel1 | 0.137 | kctd12.2 | -0.064 |
| lsp1 | 0.137 | gng2 | -0.063 |
| rhag | 0.135 | acta2 | -0.062 |
| si:ch73-68b22.2 | 0.135 | csrp1b | -0.061 |
| gata6 | 0.135 | myl9a | -0.060 |
| aqp1a.1 | 0.135 | tagln | -0.059 |
| bcam | 0.135 | cnn1b | -0.059 |
| LOC110439967 | 0.134 | cald1b | -0.058 |
| sfrp5 | 0.134 | pkig | -0.056 |
| lurap1 | 0.134 | foxf2a | -0.055 |
| CU695073.1 | 0.134 | hoxc9a | -0.055 |
| twist1b | 0.133 | hic1 | -0.055 |
| hspb1 | 0.131 | isl2b | -0.054 |
| si:ch211-117c9.1 | 0.130 | ncaldb | -0.054 |
| jam2b | 0.129 | myocd | -0.054 |
| cyp2k16 | 0.129 | LO018340.1 | -0.053 |