Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 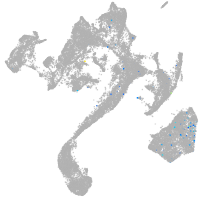 |
mural cells / non-skeletal muscle 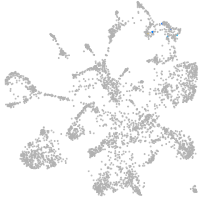 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 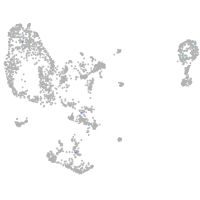 |
neural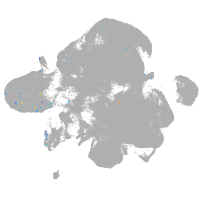 |
spinal cord / glia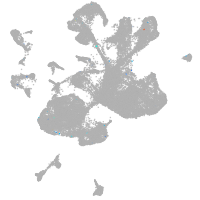 |
eye |
taste / olfactory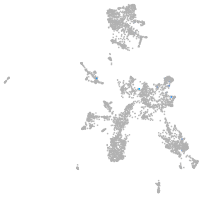 |
otic / lateral line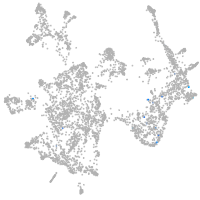 |
epidermis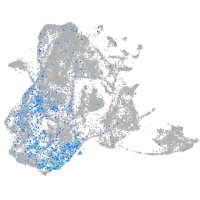 |
periderm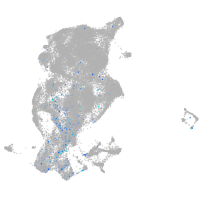 |
fin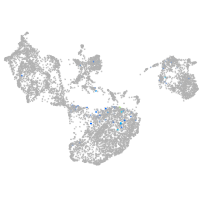 |
ionocytes / mucous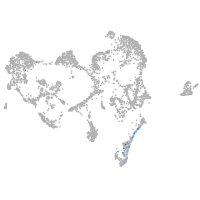 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cyp2aa3 | 0.212 | tuba1c | -0.056 |
| sdr16c5a | 0.177 | nova2 | -0.053 |
| myh9a | 0.174 | gpm6aa | -0.051 |
| cldni | 0.172 | ckbb | -0.049 |
| fermt1 | 0.171 | rtn1a | -0.045 |
| col7a1 | 0.166 | stmn1b | -0.045 |
| zgc:171775 | 0.164 | hmgb1b | -0.044 |
| padi2 | 0.163 | mdkb | -0.043 |
| spaca4l | 0.163 | rnasekb | -0.043 |
| zgc:101810 | 0.162 | tuba1a | -0.043 |
| tp63 | 0.159 | elavl3 | -0.042 |
| si:dkey-102c8.3 | 0.158 | si:ch211-137a8.4 | -0.041 |
| mir205 | 0.155 | si:dkey-276j7.1 | -0.041 |
| ecrg4b | 0.153 | atp6v0cb | -0.040 |
| capn8 | 0.151 | gpm6ab | -0.040 |
| thy1 | 0.151 | fabp3 | -0.039 |
| gpr31 | 0.150 | gng3 | -0.038 |
| cldn1 | 0.149 | pvalb1 | -0.038 |
| dnase1l4.1 | 0.148 | si:dkey-56m19.5 | -0.038 |
| tagln2 | 0.142 | hbae3 | -0.037 |
| pdlim1 | 0.140 | hbbe1.3 | -0.037 |
| pfn1 | 0.140 | pvalb2 | -0.037 |
| ptk2bb | 0.140 | cspg5a | -0.036 |
| arhgef25b | 0.139 | sncb | -0.036 |
| abi3b | 0.138 | tmsb | -0.036 |
| mapk12a | 0.138 | marcksl1a | -0.036 |
| sult2st1 | 0.138 | epb41a | -0.035 |
| lgals3b | 0.137 | vamp2 | -0.035 |
| lrata | 0.136 | tubb5 | -0.034 |
| cx44.2 | 0.132 | zc4h2 | -0.034 |
| cfl1l | 0.131 | prdx2 | -0.033 |
| itgb1b.1 | 0.131 | tox | -0.033 |
| mmp30 | 0.130 | dpysl2b | -0.033 |
| pak2a | 0.130 | actc1b | -0.032 |
| scinla | 0.130 | atp6v1e1b | -0.032 |




