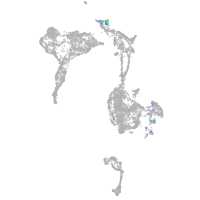
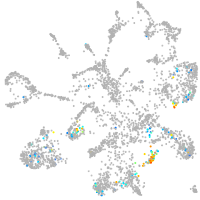
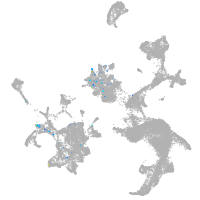
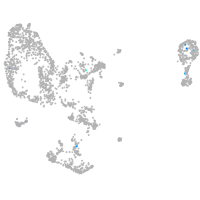
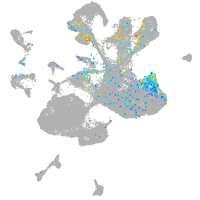
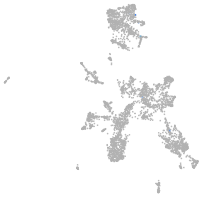
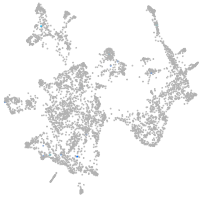
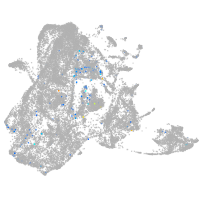
homeobox A13b
ZFIN 
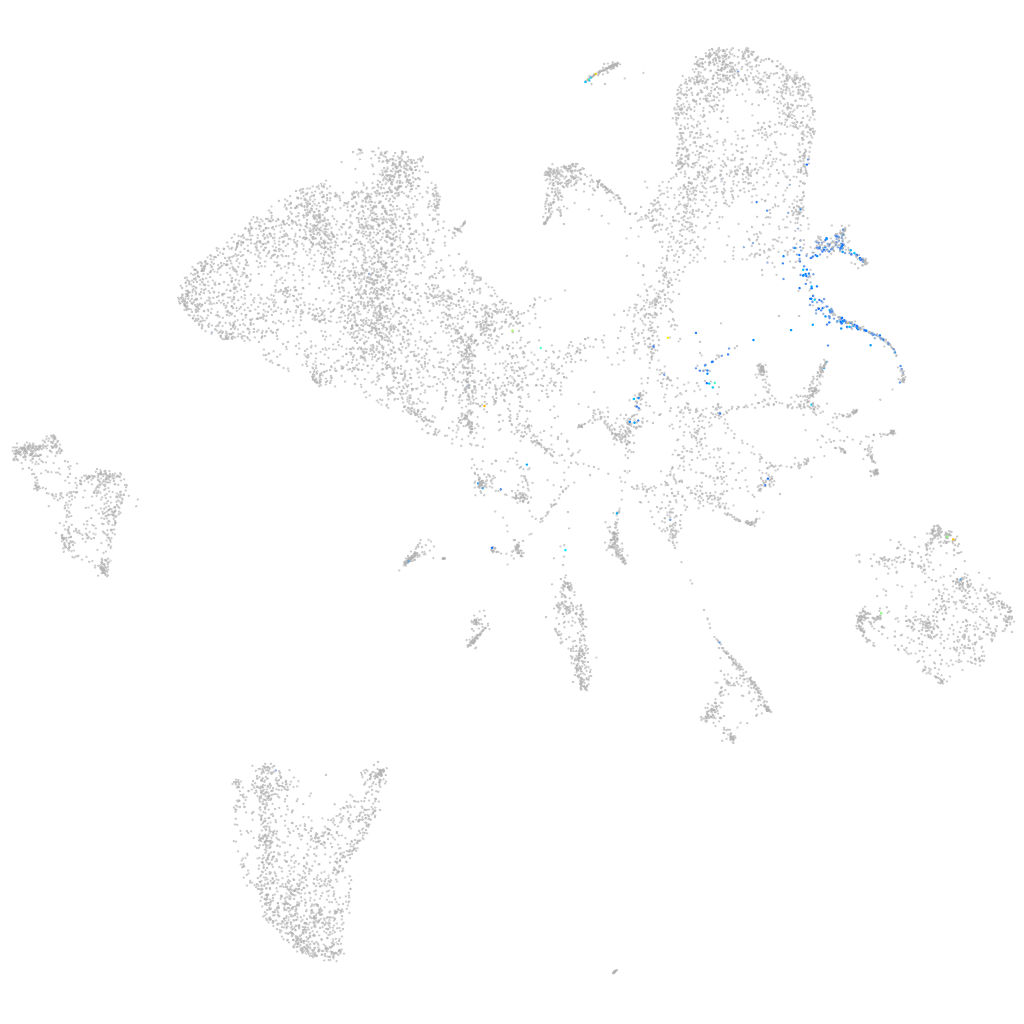
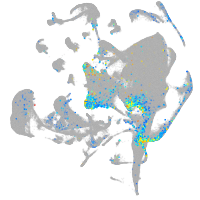
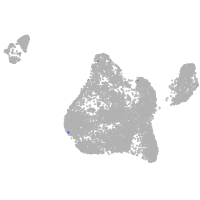
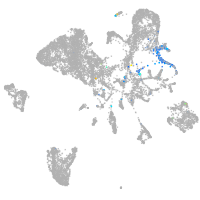
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stk25a | 0.360 | gatm | -0.122 |
| hoxa13a | 0.358 | bhmt | -0.113 |
| hoxb13a | 0.350 | aqp12 | -0.111 |
| hoxc13a | 0.346 | gamt | -0.111 |
| foxd2 | 0.326 | mat1a | -0.110 |
| ponzr1 | 0.324 | apoc2 | -0.107 |
| trim35-12 | 0.306 | si:dkey-16p21.8 | -0.099 |
| slc10a2 | 0.286 | grhprb | -0.096 |
| si:dkey-28n18.9 | 0.266 | apoa1b | -0.096 |
| sstr5 | 0.265 | nupr1b | -0.096 |
| si:ch211-202h22.9 | 0.261 | agxtb | -0.095 |
| tspan34 | 0.253 | kng1 | -0.095 |
| amn | 0.251 | cdo1 | -0.094 |
| slc12a10.1 | 0.237 | apoa2 | -0.093 |
| si:ch211-139a5.9 | 0.234 | pnp4b | -0.093 |
| si:dkey-283b1.6 | 0.234 | tfa | -0.092 |
| prr15la | 0.232 | mgst1.2 | -0.091 |
| evx1 | 0.232 | serpina1 | -0.089 |
| b3gnt3.4 | 0.229 | apobb.1 | -0.089 |
| hoxd12a | 0.227 | vtnb | -0.089 |
| dab2 | 0.224 | serpinf2b | -0.088 |
| cdaa | 0.223 | serpina1l | -0.088 |
| tmigd1 | 0.221 | apoc1 | -0.088 |
| satb2 | 0.219 | zgc:123103 | -0.086 |
| sptbn5 | 0.213 | hao1 | -0.086 |
| naga | 0.205 | fgg | -0.085 |
| hbegfa | 0.204 | ambp | -0.085 |
| si:dkey-194e6.1 | 0.203 | pla2g12b | -0.085 |
| lrp2b | 0.203 | gcshb | -0.085 |
| SPATA1 | 0.202 | apom | -0.085 |
| BX569801.1 | 0.202 | fabp10a | -0.085 |
| si:ch211-262i1.3 | 0.202 | igfbp2a | -0.085 |
| rab32b | 0.200 | uox | -0.084 |
| ctsl.1 | 0.198 | hpda | -0.084 |
| cubn | 0.197 | fgb | -0.084 |