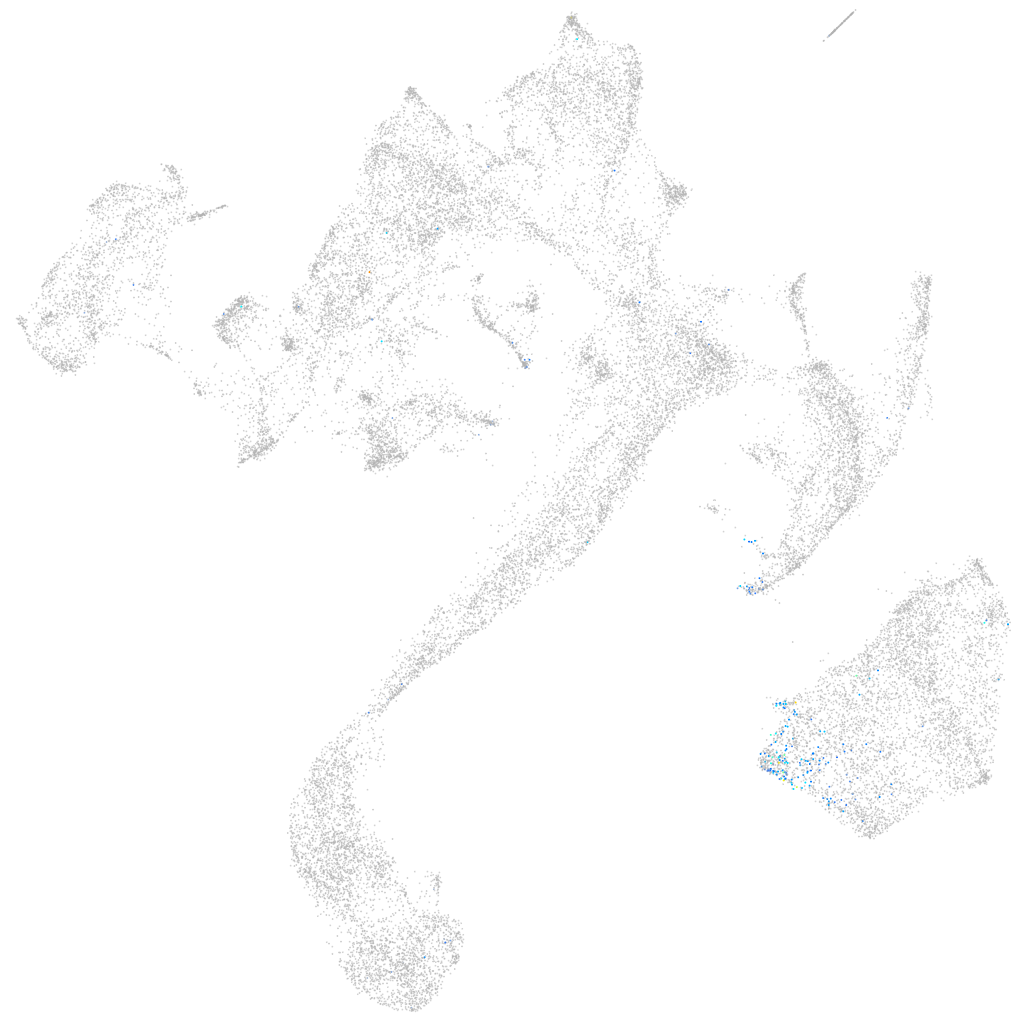
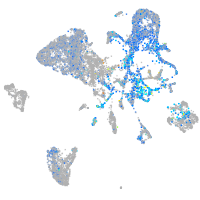
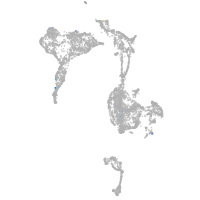
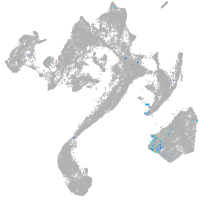
HNF1 homeobox Ba
ZFIN 
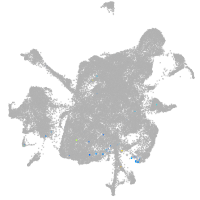
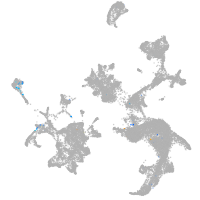
Other cell groups


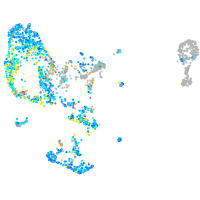
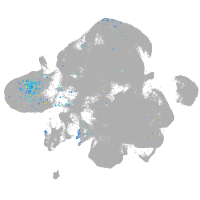
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| olig4 | 0.238 | rpl37 | -0.107 |
| her3 | 0.189 | zgc:114188 | -0.106 |
| sox19a | 0.186 | rps10 | -0.103 |
| sox3 | 0.164 | rps17 | -0.097 |
| cfhl2 | 0.159 | eef1b2 | -0.071 |
| msx1b | 0.149 | cct2 | -0.069 |
| mdkb | 0.149 | vdac2 | -0.068 |
| neurog1 | 0.147 | fabp3 | -0.065 |
| sp8b | 0.139 | zgc:56493 | -0.065 |
| alcamb | 0.133 | hnrnpa0l | -0.065 |
| chrd | 0.133 | rps23 | -0.063 |
| zic2b | 0.132 | rpl28 | -0.061 |
| ptgdsb.2 | 0.128 | rps14 | -0.060 |
| irx1b | 0.128 | rps27.1 | -0.059 |
| si:ch211-152c2.3 | 0.128 | si:ch1073-429i10.3.1 | -0.059 |
| hyal2b | 0.127 | rps25 | -0.058 |
| hoxb1b | 0.124 | vdac3 | -0.058 |
| si:ch211-137a8.4 | 0.123 | rpl9 | -0.058 |
| mir203b | 0.121 | cox4i1 | -0.057 |
| sp5l | 0.118 | mt-nd1 | -0.057 |
| noto | 0.114 | idh2 | -0.057 |
| cdx4 | 0.112 | atp5mc3b | -0.056 |
| ptgdsb.1 | 0.111 | zgc:158463 | -0.056 |
| gabrp | 0.109 | eef1g | -0.056 |
| sox2 | 0.109 | suclg1 | -0.056 |
| apela | 0.108 | rplp0 | -0.056 |
| sox19b | 0.100 | rpl23 | -0.056 |
| tle2a | 0.099 | eef1da | -0.056 |
| BX296526.2 | 0.098 | rpl19 | -0.055 |
| trim35-21 | 0.096 | rpl17 | -0.054 |
| arl4ab | 0.096 | rpl13 | -0.054 |
| nr6a1a | 0.095 | ndufa3 | -0.054 |
| hnrnpub | 0.094 | eef1db | -0.054 |
| NC-002333.4 | 0.094 | eif4a1b | -0.054 |
| ppig | 0.094 | rpl36a | -0.053 |