hemicentin 2
ZFIN 
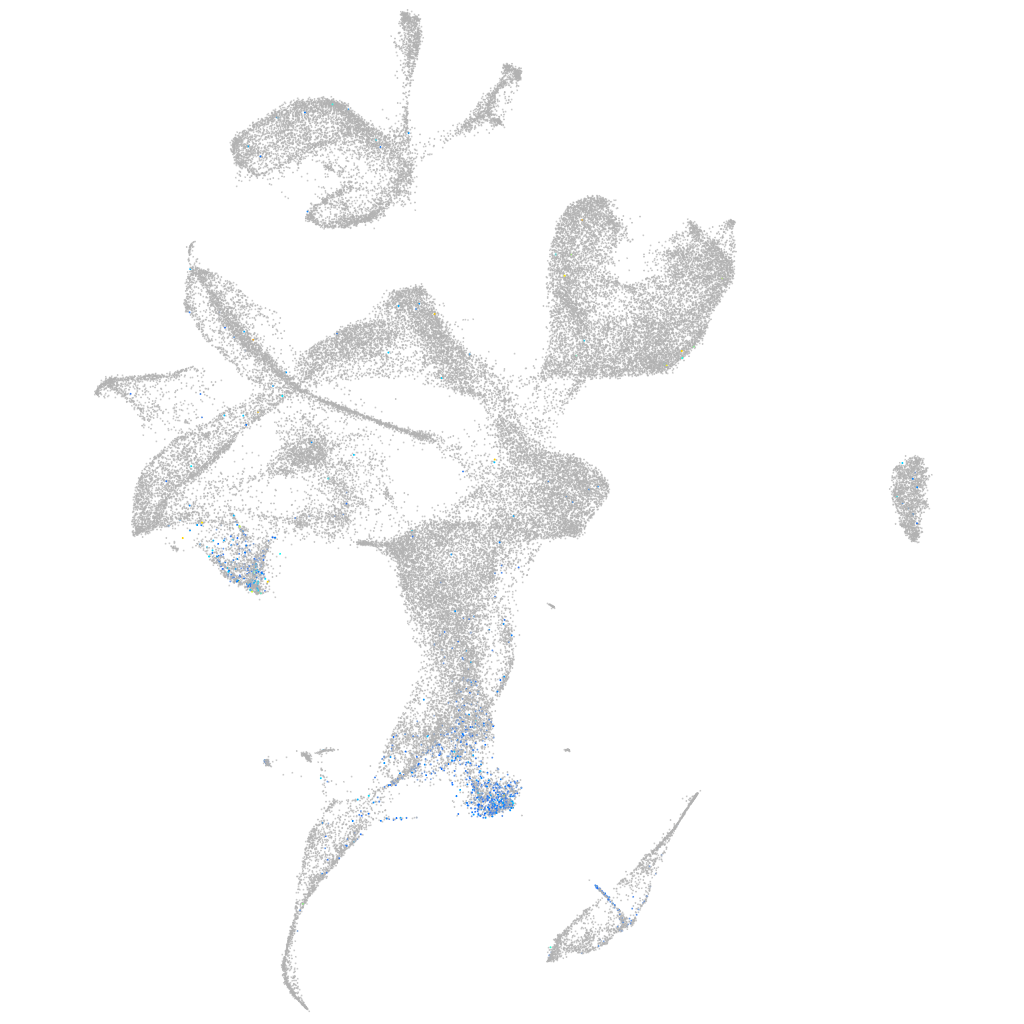
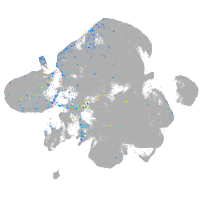
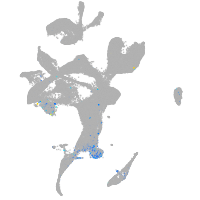
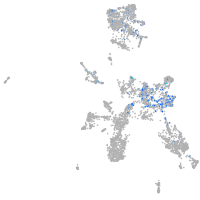
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01075068.1 | 0.203 | ckbb | -0.085 |
| eif4ebp3l | 0.163 | ptmab | -0.078 |
| cad | 0.161 | gpm6aa | -0.074 |
| bmpr1ba | 0.160 | gpm6ab | -0.074 |
| zfp36l1a | 0.157 | cadm3 | -0.070 |
| zbtb16a | 0.151 | pvalb1 | -0.067 |
| mfap2 | 0.151 | pvalb2 | -0.065 |
| npm1a | 0.146 | tuba1c | -0.065 |
| paics | 0.145 | ptmaa | -0.063 |
| CU929070.1 | 0.144 | rnasekb | -0.062 |
| si:dkey-102m7.3 | 0.142 | hmgb1a | -0.057 |
| her9 | 0.142 | actc1b | -0.055 |
| nhp2 | 0.138 | gapdhs | -0.054 |
| prps1a | 0.137 | hbbe1.3 | -0.053 |
| gart | 0.136 | rtn1a | -0.052 |
| mab21l2 | 0.135 | neurod4 | -0.051 |
| id1 | 0.135 | atp6v0cb | -0.051 |
| nip7 | 0.135 | scrt2 | -0.051 |
| mrto4 | 0.135 | fabp7a | -0.051 |
| lin28a | 0.133 | stmn1b | -0.050 |
| snu13b | 0.133 | snap25b | -0.050 |
| pprc1 | 0.132 | btg1 | -0.049 |
| apela | 0.132 | hmgn6 | -0.049 |
| lamb1a | 0.131 | ndrg4 | -0.049 |
| rcl1 | 0.129 | myt1a | -0.048 |
| vcana | 0.129 | atpv0e2 | -0.047 |
| nop58 | 0.128 | atp6v1e1b | -0.047 |
| pdcd11 | 0.128 | epb41a | -0.046 |
| aldob | 0.128 | foxg1b | -0.046 |
| fosab | 0.128 | cspg5a | -0.046 |
| dkc1 | 0.127 | hbae3 | -0.046 |
| fgfr1b | 0.127 | atp6v1g1 | -0.046 |
| slc38a5b | 0.127 | COX3 | -0.046 |
| cdkn1a | 0.127 | pdcl | -0.045 |
| HACD4 | 0.126 | h3f3c | -0.044 |