glycophorin C (Gerbich blood group)
ZFIN 
Other cell groups


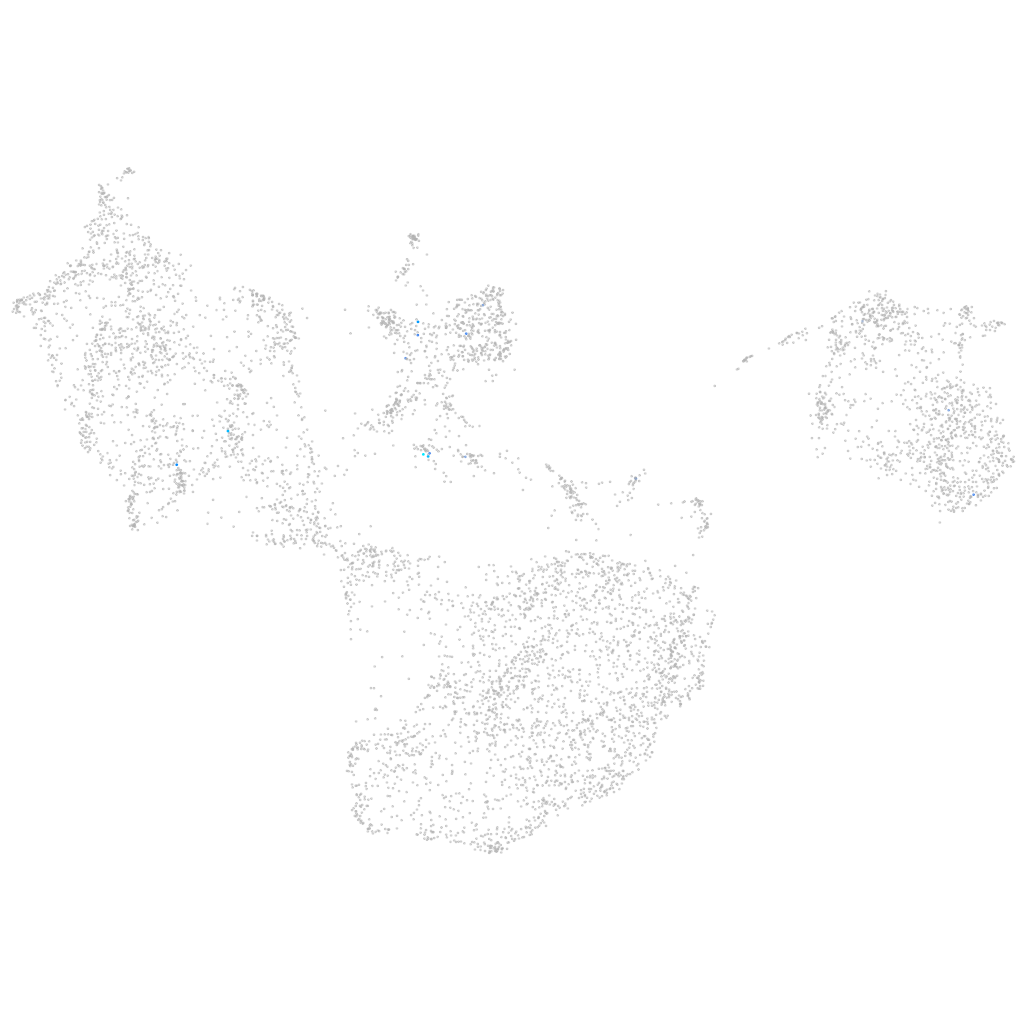
all cells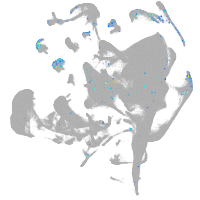 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 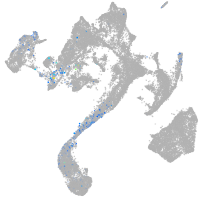 |
mural cells / non-skeletal muscle  |
mesenchyme 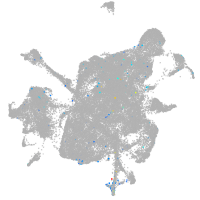 |
hematopoietic / vasculature 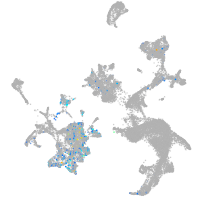 |
pronephros 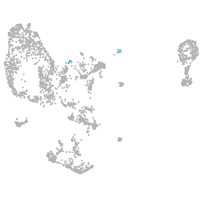 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line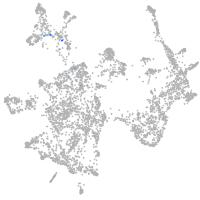 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rasip1 | 0.511 | ckap4 | -0.030 |
| lyve1a | 0.508 | XLOC-035413 | -0.028 |
| angpt2a | 0.485 | pak2a | -0.028 |
| fgd5a | 0.419 | actn4 | -0.027 |
| clec14a | 0.412 | zgc:86896 | -0.027 |
| si:ch211-33e4.2 | 0.411 | kdelr3 | -0.026 |
| LOC103909874 | 0.409 | tmsb1 | -0.026 |
| LOC101886403 | 0.394 | snd1 | -0.026 |
| XLOC-043370 | 0.394 | wbp2nl | -0.026 |
| tnfaip2b | 0.380 | cfl1l | -0.026 |
| notchl | 0.380 | cotl1 | -0.026 |
| myct1a | 0.373 | zgc:158343 | -0.025 |
| ca4b | 0.348 | cyt1 | -0.025 |
| pecam1 | 0.344 | tdh | -0.025 |
| dysf | 0.342 | GCA | -0.025 |
| LO017802.1 | 0.341 | zgc:92380 | -0.025 |
| gja5a | 0.330 | pfn1 | -0.024 |
| kdr | 0.322 | prdx6 | -0.024 |
| CR848032.1 | 0.313 | s100a10b | -0.024 |
| ecscr | 0.310 | cldn1 | -0.024 |
| acyp2 | 0.309 | tuba8l | -0.024 |
| si:dkey-42i9.16 | 0.301 | krt4 | -0.024 |
| ihha | 0.290 | cebpd | -0.024 |
| CT033796.1 | 0.266 | polr2eb | -0.023 |
| hlx1 | 0.261 | wdr1 | -0.023 |
| stab1 | 0.253 | degs1 | -0.023 |
| plvapb | 0.248 | rtn4a | -0.023 |
| tie1 | 0.246 | cd151l | -0.023 |
| mrc1a | 0.241 | arhgef5 | -0.022 |
| myct1b | 0.237 | cldni | -0.022 |
| vwf | 0.234 | mgst3a | -0.022 |
| mfng | 0.229 | zgc:153867 | -0.022 |
| LOC101885919 | 0.227 | ppp3ccb | -0.022 |
| scarf1 | 0.225 | anxa1a | -0.022 |
| cldn5b | 0.224 | tmem18 | -0.022 |