"glutathione S-transferase mu, tandem duplicate 1"
ZFIN 
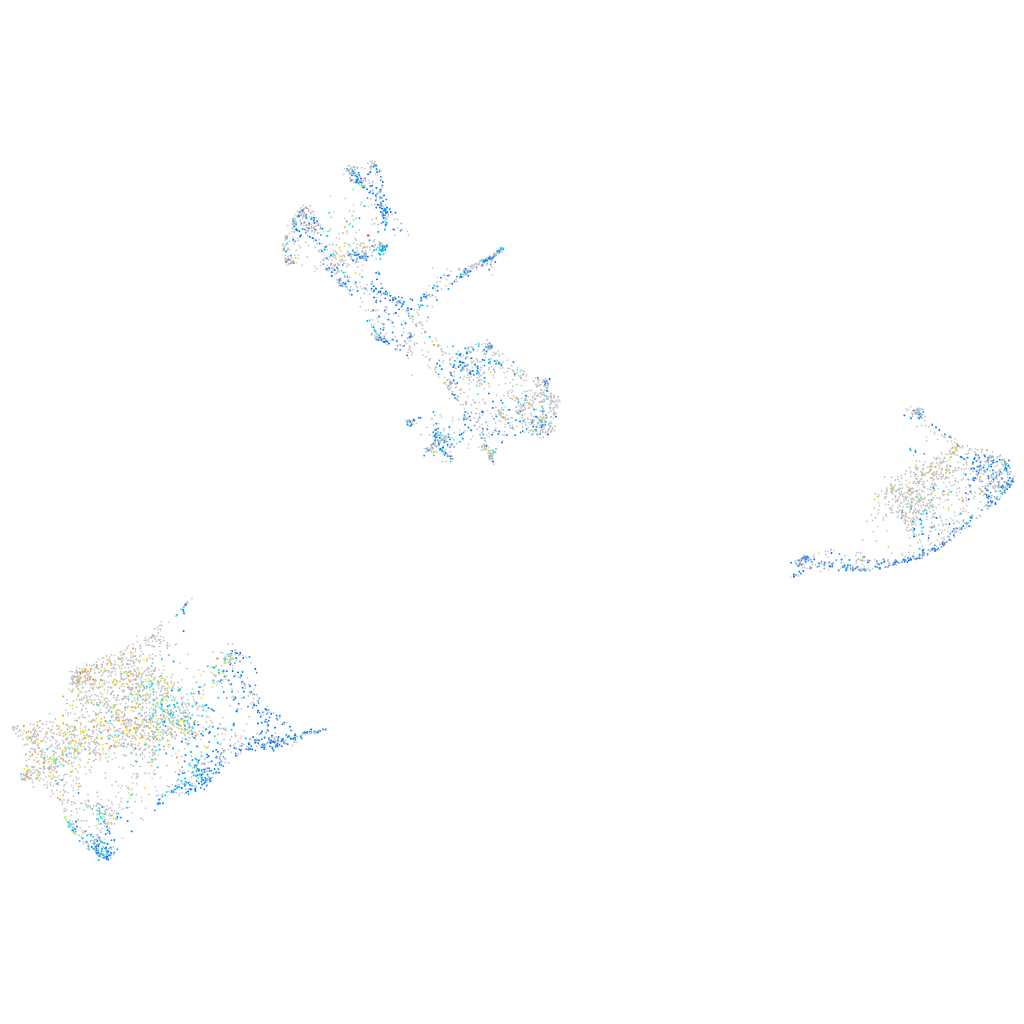
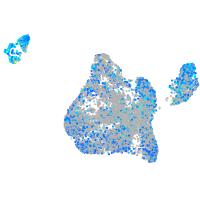
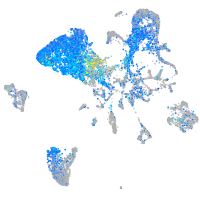
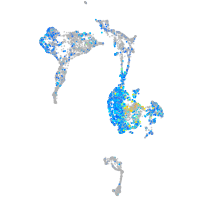
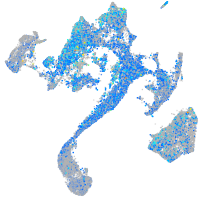
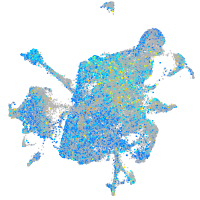
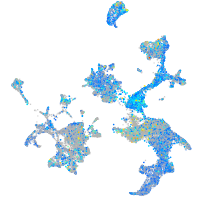
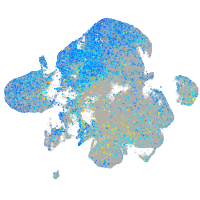
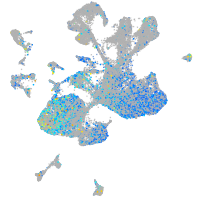
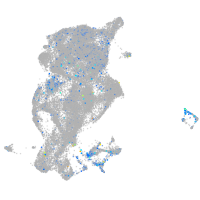
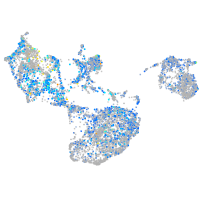
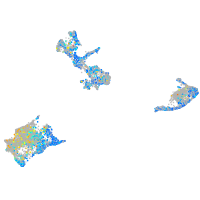
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| paics | 0.131 | tyrp1b | -0.078 |
| shmt1 | 0.123 | pmela | -0.076 |
| mdh1aa | 0.123 | zgc:91968 | -0.072 |
| iah1 | 0.122 | tyrp1a | -0.071 |
| aldob | 0.118 | dct | -0.071 |
| sod1 | 0.117 | tuba1c | -0.068 |
| phyhd1 | 0.114 | elavl3 | -0.068 |
| atic | 0.111 | prkar1b | -0.062 |
| sigmar1 | 0.110 | oca2 | -0.062 |
| impdh1b | 0.108 | gpm6aa | -0.060 |
| akr1b1 | 0.108 | mtbl | -0.060 |
| sult1st1 | 0.108 | gng3 | -0.057 |
| prdx5 | 0.107 | nectin1b | -0.056 |
| cyb5a | 0.107 | nova2 | -0.056 |
| prps1a | 0.106 | gap43 | -0.055 |
| gart | 0.106 | myt1b | -0.054 |
| uraha | 0.105 | myt1a | -0.054 |
| chp2 | 0.105 | tuba1a | -0.053 |
| aldh1l1 | 0.103 | hbae1.1 | -0.052 |
| ppat | 0.102 | tmeff1b | -0.052 |
| si:dkey-251i10.2 | 0.102 | LOC100537384 | -0.052 |
| glrx | 0.101 | zc4h2 | -0.052 |
| coro1a | 0.101 | ptmaa | -0.051 |
| mibp | 0.100 | elavl4 | -0.050 |
| aox5 | 0.099 | rtn1a | -0.050 |
| glulb | 0.099 | tnni2a.4 | -0.050 |
| prdx6 | 0.098 | hbbe1.3 | -0.049 |
| pon1 | 0.098 | ptmab | -0.049 |
| gstp1 | 0.098 | stmn1b | -0.048 |
| prtfdc1 | 0.097 | cbfa2t3 | -0.048 |
| krt18b | 0.097 | celf3a | -0.048 |
| alas1 | 0.096 | mapk10 | -0.047 |
| fxyd6l | 0.096 | cadm3 | -0.047 |
| gmps | 0.095 | dpysl5a | -0.047 |
| prdx1 | 0.095 | fez1 | -0.046 |