growth factor receptor-bound protein 2b
ZFIN 
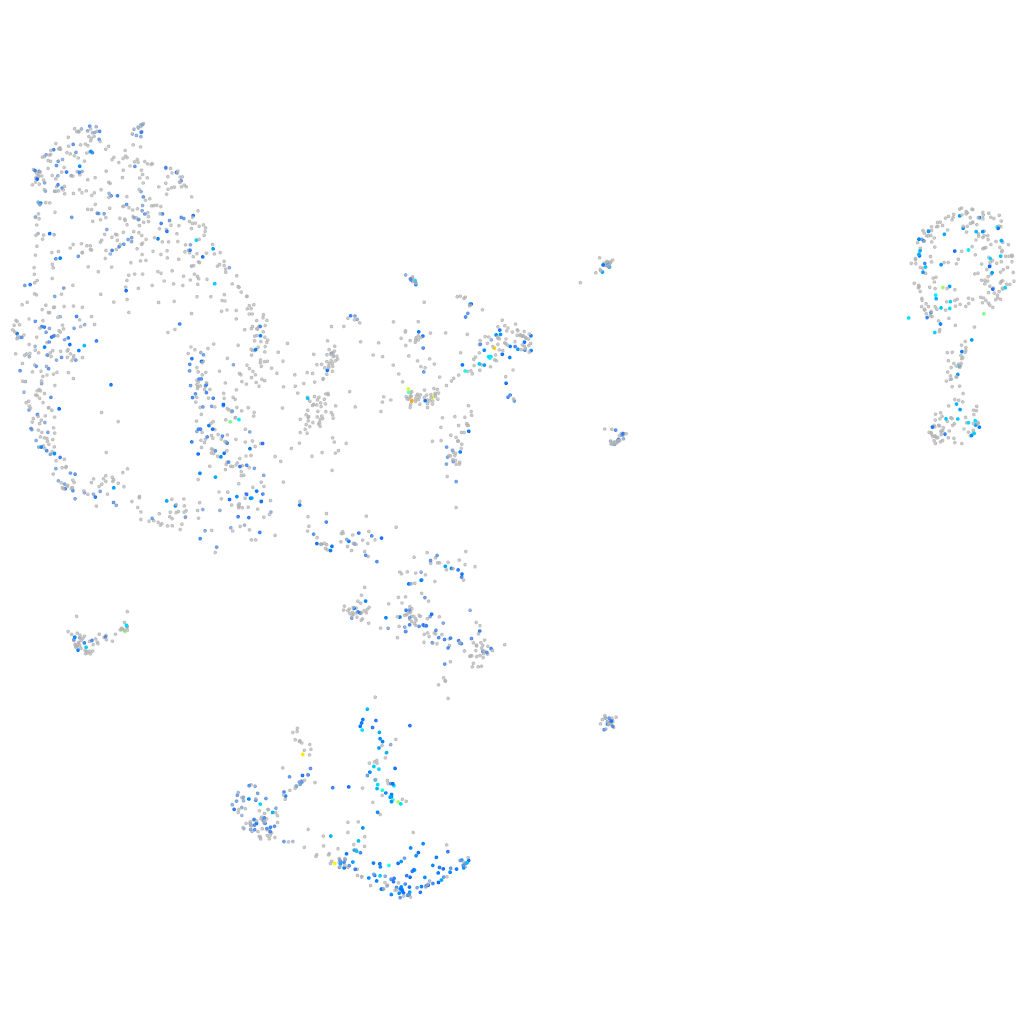
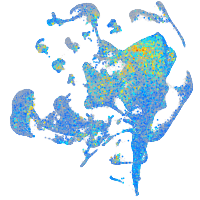
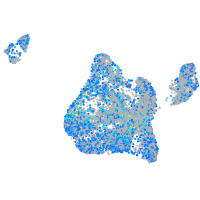
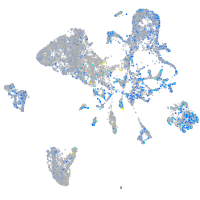
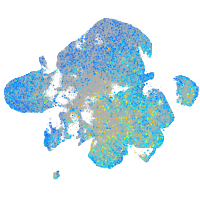
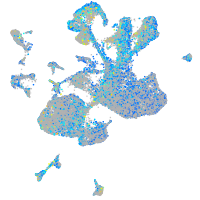
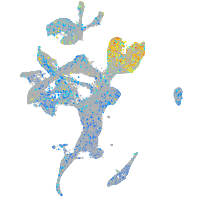
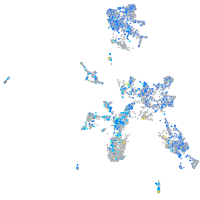
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp1a1a.3 | 0.284 | grhprb | -0.156 |
| rnf183 | 0.269 | acy3.2 | -0.156 |
| si:dkey-36i7.3 | 0.267 | pdzk1ip1 | -0.146 |
| si:ch73-359m17.9 | 0.263 | gstt1a | -0.141 |
| trim35-12 | 0.259 | gstr | -0.140 |
| bsnd | 0.251 | dab2 | -0.140 |
| gata3 | 0.249 | fbp1b | -0.139 |
| soul2 | 0.244 | pnp6 | -0.139 |
| phlda2 | 0.241 | nit2 | -0.138 |
| agr2 | 0.238 | si:dkey-194e6.1 | -0.137 |
| ppp1r1b | 0.236 | upb1 | -0.133 |
| zgc:158423 | 0.234 | lrp2a | -0.131 |
| AL954322.2 | 0.227 | si:dkey-33i11.9 | -0.131 |
| oclnb | 0.224 | alpl | -0.131 |
| ccl19a.1 | 0.222 | pdzk1 | -0.129 |
| tbx2b | 0.219 | gpt2l | -0.128 |
| clcnk | 0.219 | hao1 | -0.128 |
| mctp2b | 0.219 | aldob | -0.128 |
| tmem72 | 0.217 | pklr | -0.128 |
| ca4c | 0.217 | slc26a1 | -0.127 |
| degs2 | 0.216 | cubn | -0.126 |
| mal2 | 0.216 | zgc:136493 | -0.126 |
| tmem176l.4 | 0.215 | gamt | -0.126 |
| gdpd3a | 0.214 | aqp8b | -0.125 |
| prkag2a | 0.213 | cx32.3 | -0.124 |
| malb | 0.211 | agxtb | -0.124 |
| gpa33a | 0.211 | agxta | -0.122 |
| slc12a3 | 0.210 | hoga1 | -0.122 |
| cldne | 0.207 | slc20a1a | -0.121 |
| hoxb8a | 0.207 | dpydb | -0.120 |
| etf1a | 0.205 | slc23a1 | -0.119 |
| erbb3a | 0.205 | gcshb | -0.119 |
| hoxc13b | 0.203 | epdl2 | -0.118 |
| slc9a3.2 | 0.201 | xirp1 | -0.118 |
| gucy2c | 0.200 | si:dkey-151g10.6 | -0.117 |