G protein-coupled receptor 173
ZFIN 
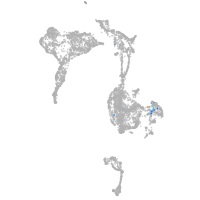
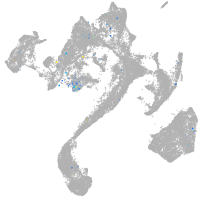
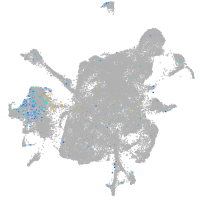
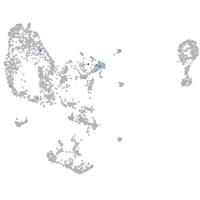
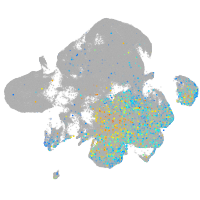
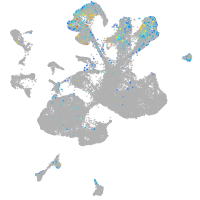
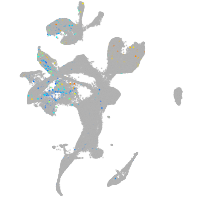
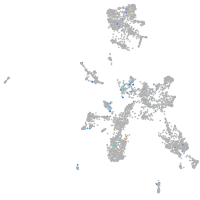
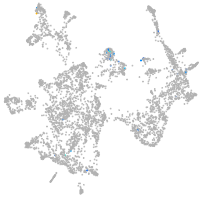
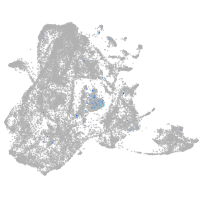
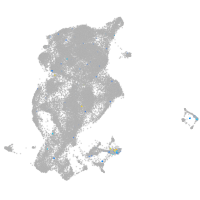
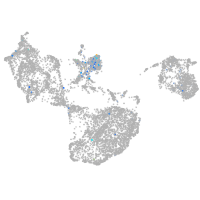
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nova1 | 0.268 | si:dkey-151g10.6 | -0.086 |
| sncb | 0.246 | tspan36 | -0.082 |
| rai1 | 0.245 | eef1a1l1 | -0.075 |
| nrxn1a | 0.244 | rpl36a | -0.074 |
| atp1a3a | 0.236 | rpl11 | -0.072 |
| gng3 | 0.235 | smim29 | -0.068 |
| vamp2 | 0.232 | rplp2l | -0.067 |
| syn2a | 0.232 | rplp0 | -0.066 |
| cadm4 | 0.225 | pfn1 | -0.066 |
| CU628002.1 | 0.222 | pcbd1 | -0.064 |
| slc6a1a | 0.222 | rplp1 | -0.062 |
| elavl4 | 0.220 | qdpra | -0.062 |
| zgc:153426 | 0.217 | akr1b1 | -0.061 |
| eno2 | 0.217 | gstp1 | -0.060 |
| stmn2a | 0.212 | rps2 | -0.059 |
| BX255967.1 | 0.212 | gpr143 | -0.058 |
| gpm6aa | 0.212 | rpl12 | -0.057 |
| RYR2 | 0.212 | pts | -0.057 |
| celf5a | 0.211 | rnaseka | -0.056 |
| BX005003.1 | 0.211 | mitfa | -0.055 |
| rnasekb | 0.209 | atic | -0.055 |
| stmn1b | 0.207 | pttg1ipb | -0.054 |
| tuba1c | 0.206 | atox1 | -0.054 |
| pcdh2ab7 | 0.205 | rps12 | -0.053 |
| snap25a | 0.202 | rpl23 | -0.053 |
| si:dkey-71h2.2 | 0.202 | rabl6b | -0.053 |
| stx1b | 0.200 | rab32a | -0.053 |
| amph | 0.199 | rpl29 | -0.053 |
| atp6v0cb | 0.199 | rpsa | -0.053 |
| clvs2 | 0.198 | rpl7 | -0.053 |
| sypb | 0.198 | rps19 | -0.053 |
| gabrb4 | 0.198 | slc38a5b | -0.052 |
| sprn | 0.197 | bace2 | -0.052 |
| sv2a | 0.197 | eef1da | -0.052 |
| nova2 | 0.197 | syngr1a | -0.052 |