GNAS complex locus
ZFIN 
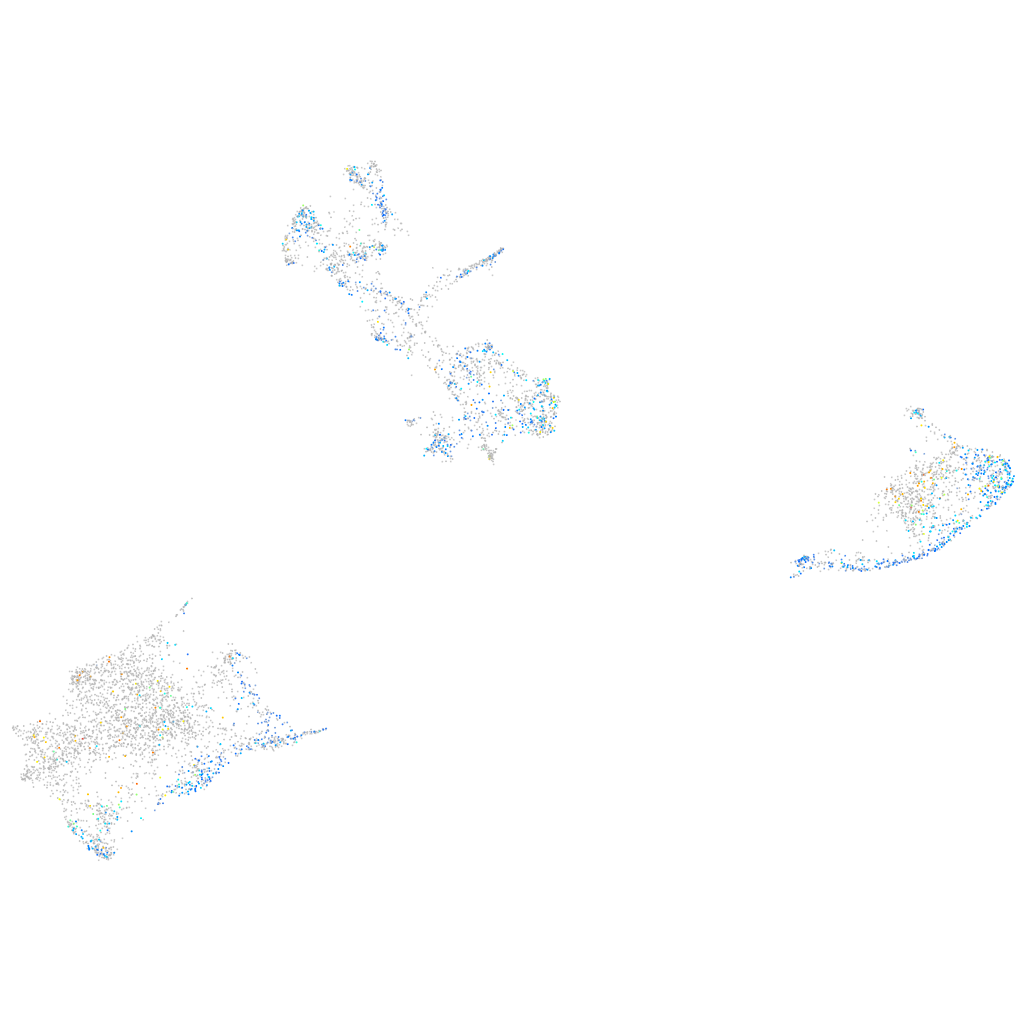
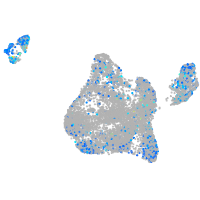
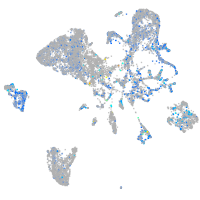
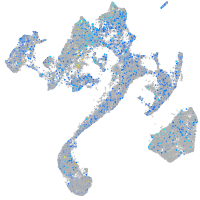
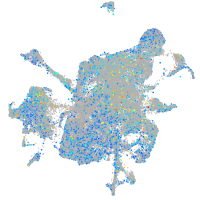
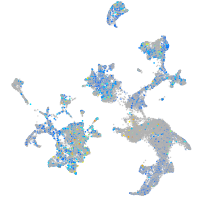
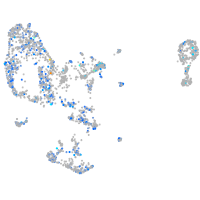
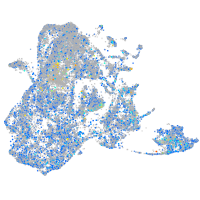
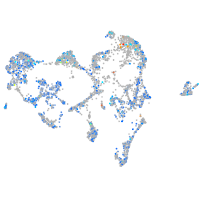
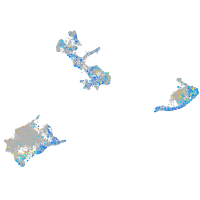
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| oca2 | 0.181 | CABZ01021592.1 | -0.130 |
| kita | 0.181 | si:dkey-251i10.2 | -0.121 |
| SPAG9 | 0.172 | paics | -0.103 |
| lamp1a | 0.163 | uraha | -0.099 |
| CDK18 | 0.159 | mdh1aa | -0.085 |
| tyrp1b | 0.159 | gpd1b | -0.082 |
| pmela | 0.158 | tmem130 | -0.080 |
| dct | 0.158 | gch2 | -0.080 |
| tyrp1a | 0.157 | hbbe1.3 | -0.078 |
| tyr | 0.157 | aldob | -0.069 |
| slc22a2 | 0.157 | cyb5a | -0.069 |
| slc24a5 | 0.156 | mylpfa | -0.067 |
| prkar1b | 0.156 | pts | -0.067 |
| spock3 | 0.149 | prdx5 | -0.065 |
| zgc:91968 | 0.148 | prdx1 | -0.065 |
| slc45a2 | 0.146 | actc1b | -0.064 |
| atp6v0ca | 0.144 | si:dkey-151g10.6 | -0.063 |
| CR383676.1 | 0.144 | hbae3 | -0.063 |
| kcnj13 | 0.142 | slc22a7a | -0.063 |
| si:ch73-389b16.1 | 0.141 | aox5 | -0.062 |
| slc39a1 | 0.140 | akr1b1 | -0.062 |
| mtbl | 0.139 | sult1st1 | -0.062 |
| slc43a2a | 0.139 | mylz3 | -0.059 |
| tfap2e | 0.138 | pvalb2 | -0.059 |
| rap1gap | 0.138 | pvalb1 | -0.057 |
| si:zfos-943e10.1 | 0.138 | atic | -0.057 |
| mchr2 | 0.136 | hbbe1.1 | -0.054 |
| CR847566.1 | 0.135 | myhz1.2 | -0.054 |
| slc7a5 | 0.135 | hbae1.1 | -0.054 |
| slc29a3 | 0.134 | TMEM19 | -0.054 |
| aadac | 0.134 | bco1 | -0.052 |
| slc39a10 | 0.133 | rbp4l | -0.051 |
| tfap2a | 0.132 | si:ch211-251b21.1 | -0.050 |
| GNAZ | 0.130 | krt4 | -0.049 |
| mlpha | 0.129 | zgc:153031 | -0.047 |