glutamate-ammonia ligase (glutamine synthase) b
ZFIN 
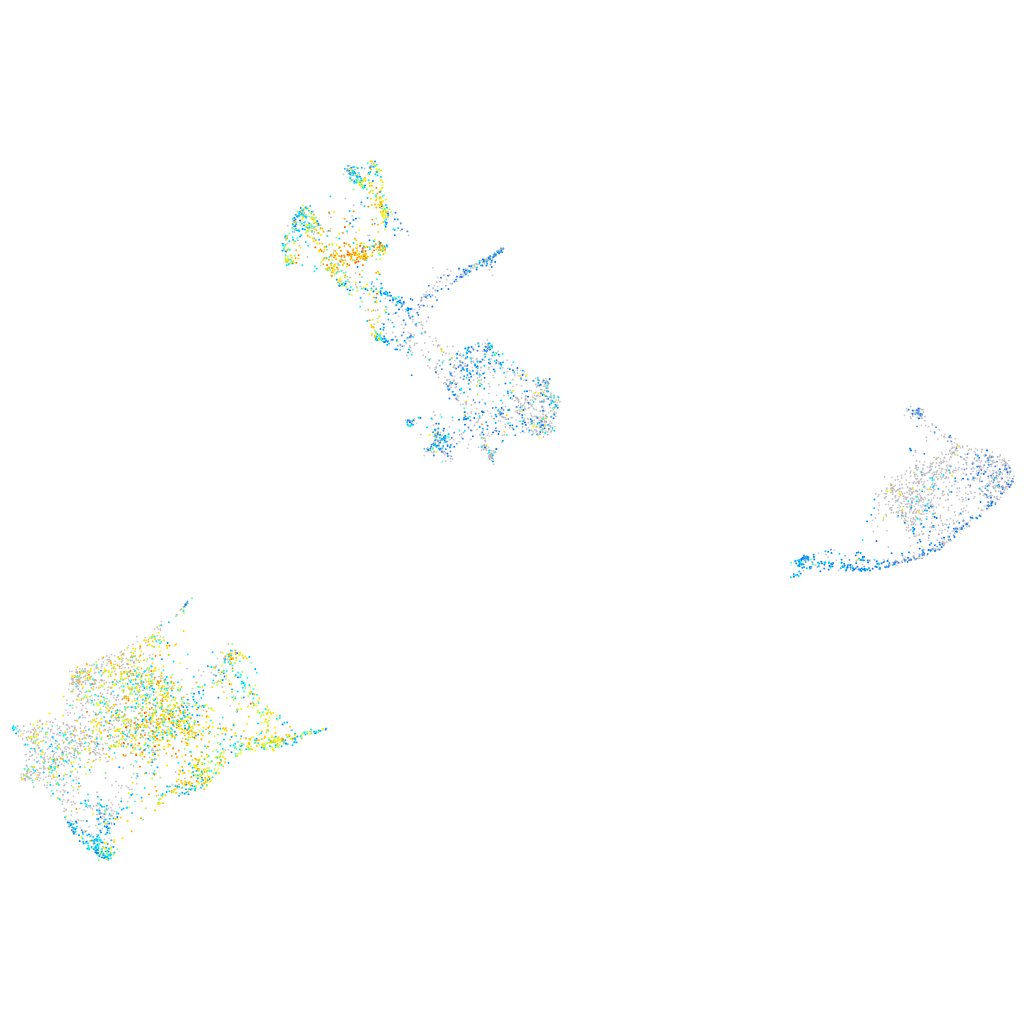
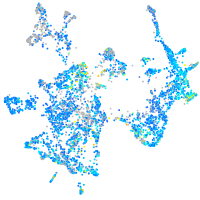
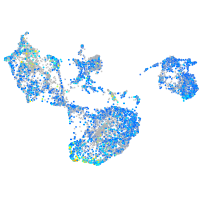
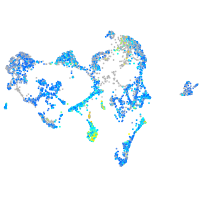
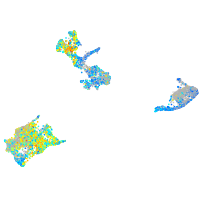
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| impdh1b | 0.540 | tyrp1b | -0.370 |
| paics | 0.492 | zgc:91968 | -0.346 |
| gmps | 0.463 | tyrp1a | -0.343 |
| atic | 0.451 | pmela | -0.343 |
| slc2a15a | 0.451 | dct | -0.334 |
| prps1a | 0.447 | oca2 | -0.315 |
| prdx1 | 0.385 | icn | -0.309 |
| mdh1aa | 0.363 | s100a10b | -0.308 |
| cyb5a | 0.361 | prkar1b | -0.290 |
| ppat | 0.355 | si:ch73-389b16.1 | -0.290 |
| gart | 0.344 | anxa1a | -0.285 |
| sprb | 0.341 | slc24a5 | -0.266 |
| akr1b1 | 0.318 | kita | -0.258 |
| uraha | 0.316 | mtbl | -0.258 |
| pltp | 0.308 | ckbb | -0.244 |
| cx30.3 | 0.305 | slc39a10 | -0.228 |
| tagln3b | 0.303 | gstt1a | -0.227 |
| adsl | 0.302 | mlpha | -0.225 |
| aox5 | 0.298 | slc22a2 | -0.222 |
| zgc:110789 | 0.296 | aadac | -0.221 |
| slc25a36a | 0.293 | spock3 | -0.218 |
| pts | 0.291 | ptmaa | -0.203 |
| akap12a | 0.283 | hsd20b2 | -0.199 |
| mibp | 0.282 | tyr | -0.194 |
| phgdh | 0.281 | slc6a15 | -0.193 |
| pgm2 | 0.281 | slc29a3 | -0.186 |
| lypc | 0.280 | zdhhc2 | -0.184 |
| slc2a11b | 0.279 | yjefn3 | -0.178 |
| psat1 | 0.276 | rap1gap | -0.176 |
| pnp4a | 0.276 | gpr61 | -0.173 |
| pfas | 0.275 | slc7a5 | -0.170 |
| sod1 | 0.274 | CDK18 | -0.170 |
| tpd52 | 0.272 | spra | -0.166 |
| coro1a | 0.272 | gramd2aa | -0.166 |
| shmt1 | 0.271 | slc30a1b | -0.165 |