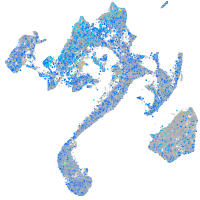
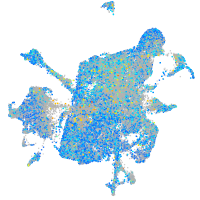
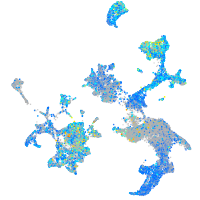
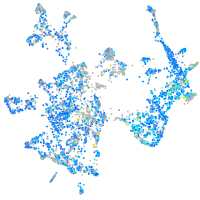
glutaredoxin (thioltransferase)
ZFIN 
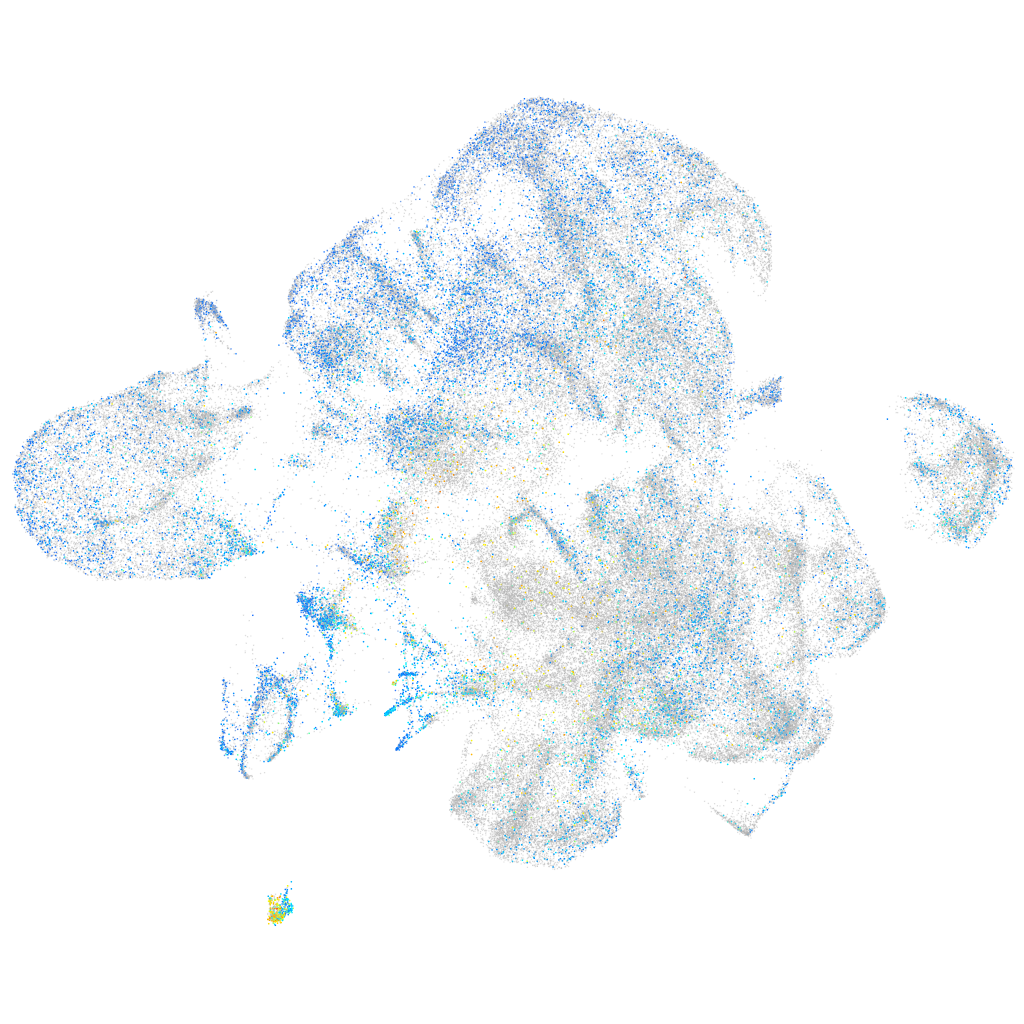
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| grp | 0.188 | nova2 | -0.101 |
| cldnh | 0.186 | gpm6aa | -0.093 |
| vill | 0.179 | elavl3 | -0.087 |
| gfral | 0.178 | tuba1c | -0.078 |
| prr15la | 0.177 | rtn1a | -0.074 |
| zgc:92380 | 0.175 | stmn1b | -0.073 |
| itpr3 | 0.173 | marcksl1b | -0.069 |
| CR759836.1 | 0.173 | gpm6ab | -0.063 |
| pou2f3 | 0.167 | hmgb3a | -0.063 |
| pvalb8 | 0.167 | ckbb | -0.063 |
| zgc:101744 | 0.166 | CU467822.1 | -0.062 |
| si:ch1073-443f11.2 | 0.163 | chd4a | -0.062 |
| zgc:123295 | 0.159 | celf2 | -0.062 |
| gcnt7 | 0.159 | mdkb | -0.062 |
| ponzr5 | 0.157 | zc4h2 | -0.060 |
| si:ch211-256e16.6 | 0.156 | tubb5 | -0.060 |
| hepacam2 | 0.155 | tuba1a | -0.058 |
| ovol1a | 0.154 | tmsb | -0.055 |
| b4galnt3b | 0.154 | si:dkey-56m19.5 | -0.055 |
| si:ch211-130m23.2 | 0.154 | ptmab | -0.055 |
| hopx | 0.153 | myt1la | -0.055 |
| plcd1a | 0.151 | scrt2 | -0.055 |
| CABZ01073083.1 | 0.150 | CU634008.1 | -0.054 |
| degs2 | 0.149 | ebf3a | -0.053 |
| bik | 0.148 | fam168a | -0.053 |
| epcam | 0.146 | scrt1a | -0.053 |
| spint2 | 0.146 | gatad2b | -0.053 |
| tmem45b | 0.144 | bcl11ba | -0.052 |
| si:ch211-202h22.8 | 0.143 | si:ch211-222l21.1 | -0.052 |
| krt8 | 0.142 | sox4a | -0.051 |
| cldnb | 0.137 | nr2f1a | -0.051 |
| si:ch211-79k12.1 | 0.136 | thsd7aa | -0.051 |
| s100u | 0.136 | pbx3b | -0.051 |
| anxa5b | 0.136 | si:dkey-276j7.1 | -0.050 |
| cers3b | 0.133 | FO082781.1 | -0.050 |