GDNF family receptor alpha 3
ZFIN 
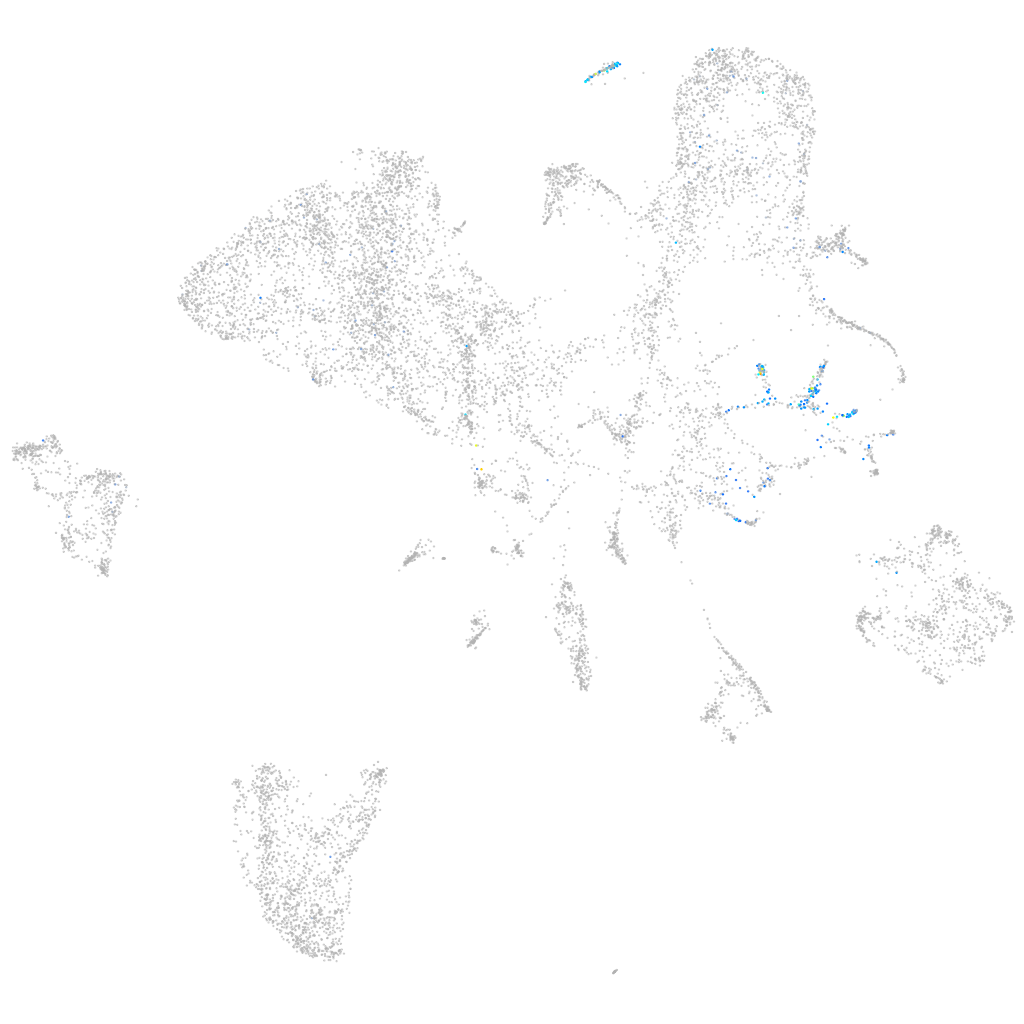
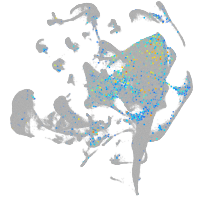
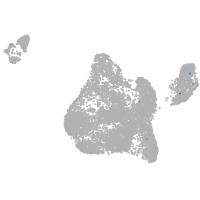
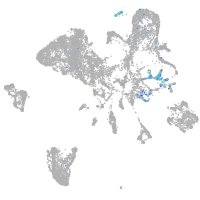
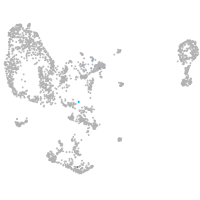
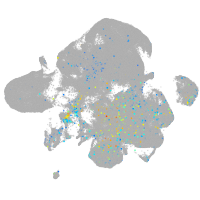
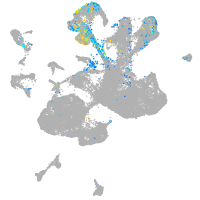
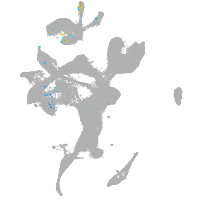
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.429 | gamt | -0.118 |
| neurod1 | 0.396 | hdlbpa | -0.117 |
| egr4 | 0.379 | ahcy | -0.115 |
| XLOC-007078 | 0.376 | gapdh | -0.115 |
| fev | 0.375 | fabp3 | -0.109 |
| pax6b | 0.355 | gatm | -0.104 |
| si:ch73-160i9.2 | 0.353 | aldh6a1 | -0.102 |
| zgc:101731 | 0.339 | bhmt | -0.101 |
| insm1b | 0.335 | fbp1b | -0.098 |
| rasd4 | 0.324 | mat1a | -0.093 |
| mir7a-1 | 0.323 | nupr1b | -0.093 |
| nkx2.2a | 0.318 | cx32.3 | -0.092 |
| c2cd4a | 0.317 | apoa1b | -0.091 |
| pcsk1nl | 0.317 | si:dkey-16p21.8 | -0.091 |
| TCIM (1 of many) | 0.313 | rpl6 | -0.091 |
| mir375-2 | 0.312 | gstt1a | -0.089 |
| pcsk1 | 0.310 | aqp12 | -0.088 |
| sprn2 | 0.303 | apoa4b.1 | -0.087 |
| vat1 | 0.293 | apoc2 | -0.087 |
| hepacam2 | 0.292 | aldh7a1 | -0.087 |
| dll4 | 0.287 | gnmt | -0.086 |
| pax4 | 0.286 | rpl4 | -0.086 |
| etv1 | 0.286 | gstr | -0.085 |
| lysmd2 | 0.286 | cebpd | -0.085 |
| syt1a | 0.285 | aldob | -0.085 |
| capsla | 0.285 | agxta | -0.085 |
| mapk15 | 0.283 | afp4 | -0.085 |
| rprmb | 0.281 | apoc1 | -0.084 |
| ccka | 0.277 | agxtb | -0.082 |
| arxa | 0.273 | mgst1.2 | -0.082 |
| gcgb | 0.273 | ppa1b | -0.081 |
| nmbb | 0.272 | apoa2 | -0.081 |
| scgn | 0.265 | wu:fj16a03 | -0.081 |
| adcyap1a | 0.263 | sec61a1 | -0.081 |
| vamp2 | 0.263 | shmt1 | -0.080 |