"gamma-aminobutyric acid (GABA) A receptor, gamma 2"
ZFIN 
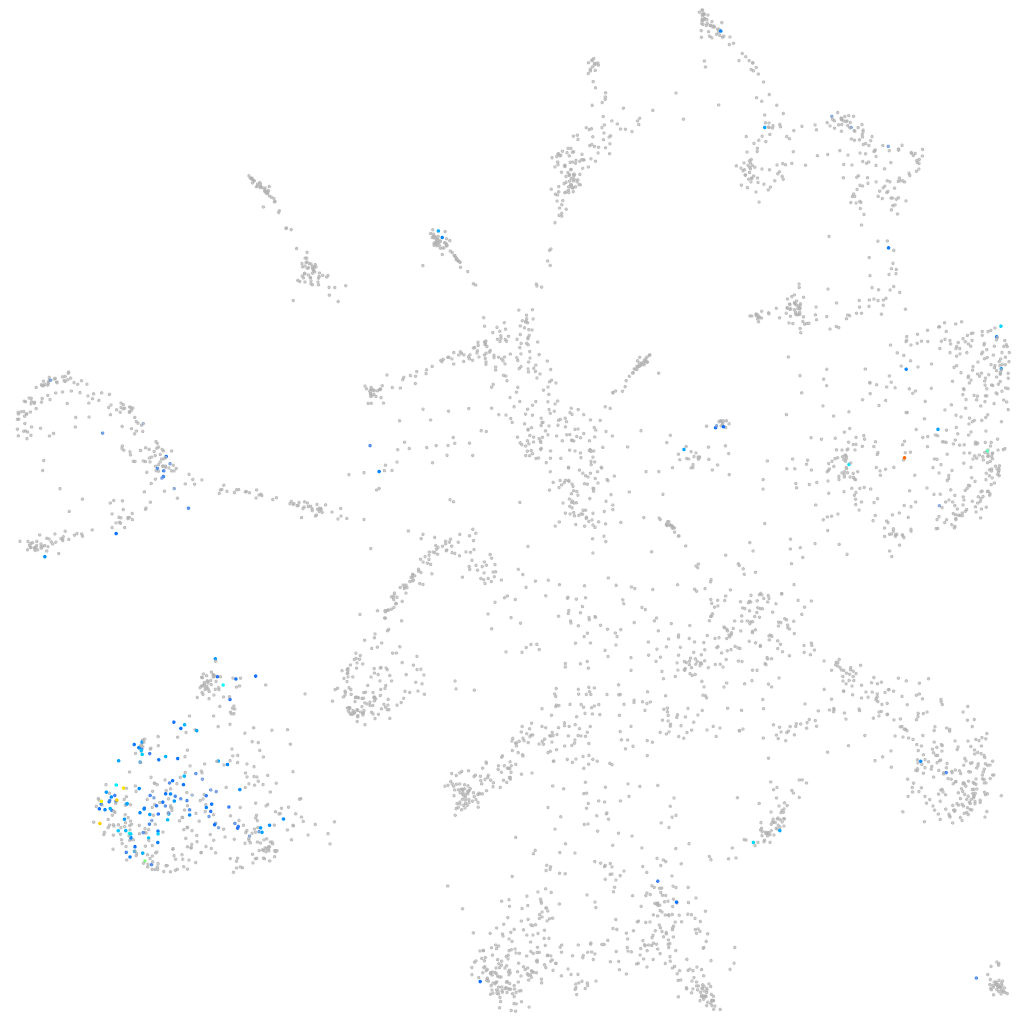
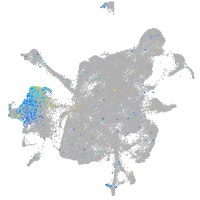
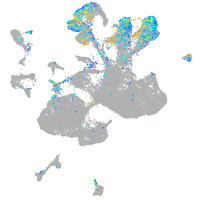
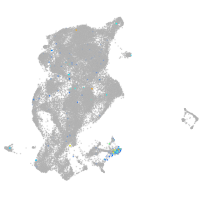
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sypa | 0.368 | rps27a | -0.082 |
| sv2a | 0.352 | fstl1b | -0.073 |
| cspg5a | 0.346 | rbpms2a | -0.068 |
| vamp2 | 0.340 | rplp1 | -0.067 |
| stx1b | 0.339 | rps8a | -0.066 |
| sncb | 0.339 | rps13 | -0.063 |
| stxbp1a | 0.339 | rplp2l | -0.061 |
| elavl4 | 0.333 | rps19 | -0.060 |
| gabra1 | 0.325 | eef1a1l1 | -0.057 |
| rtn1b | 0.325 | rpl18 | -0.056 |
| slc6a1a | 0.322 | pmp22a | -0.055 |
| snap25a | 0.321 | rpsa | -0.055 |
| syt1a | 0.317 | rps10 | -0.054 |
| gpm6ab | 0.312 | rps2 | -0.054 |
| ywhag2 | 0.311 | rpl12 | -0.052 |
| gng3 | 0.311 | rpl8 | -0.052 |
| map1aa | 0.308 | rpl27a | -0.051 |
| atp6v0cb | 0.308 | rps28 | -0.050 |
| stmn1b | 0.307 | rps11 | -0.048 |
| nmnat2 | 0.307 | rpl18a | -0.048 |
| zgc:65894 | 0.306 | rps24 | -0.047 |
| eno2 | 0.302 | actc1b | -0.047 |
| tuba1c | 0.300 | rps9 | -0.046 |
| gnao1a | 0.299 | BX901920.1 | -0.045 |
| gabrb4 | 0.297 | rpl26 | -0.045 |
| stmn2a | 0.295 | rps4x | -0.045 |
| nova2 | 0.294 | XLOC-028370 | -0.045 |
| rtn1a | 0.293 | rpl27 | -0.045 |
| mllt11 | 0.288 | tpt1 | -0.044 |
| elavl3 | 0.288 | rpl14 | -0.044 |
| ube2ql1 | 0.283 | rpl7a | -0.043 |
| myt1la | 0.281 | rpl28 | -0.043 |
| nova1 | 0.280 | zgc:162730 | -0.042 |
| syn2a | 0.277 | rpl24 | -0.042 |
| gpm6aa | 0.275 | rps16 | -0.042 |