GA binding protein transcription factor subunit alpha
ZFIN 
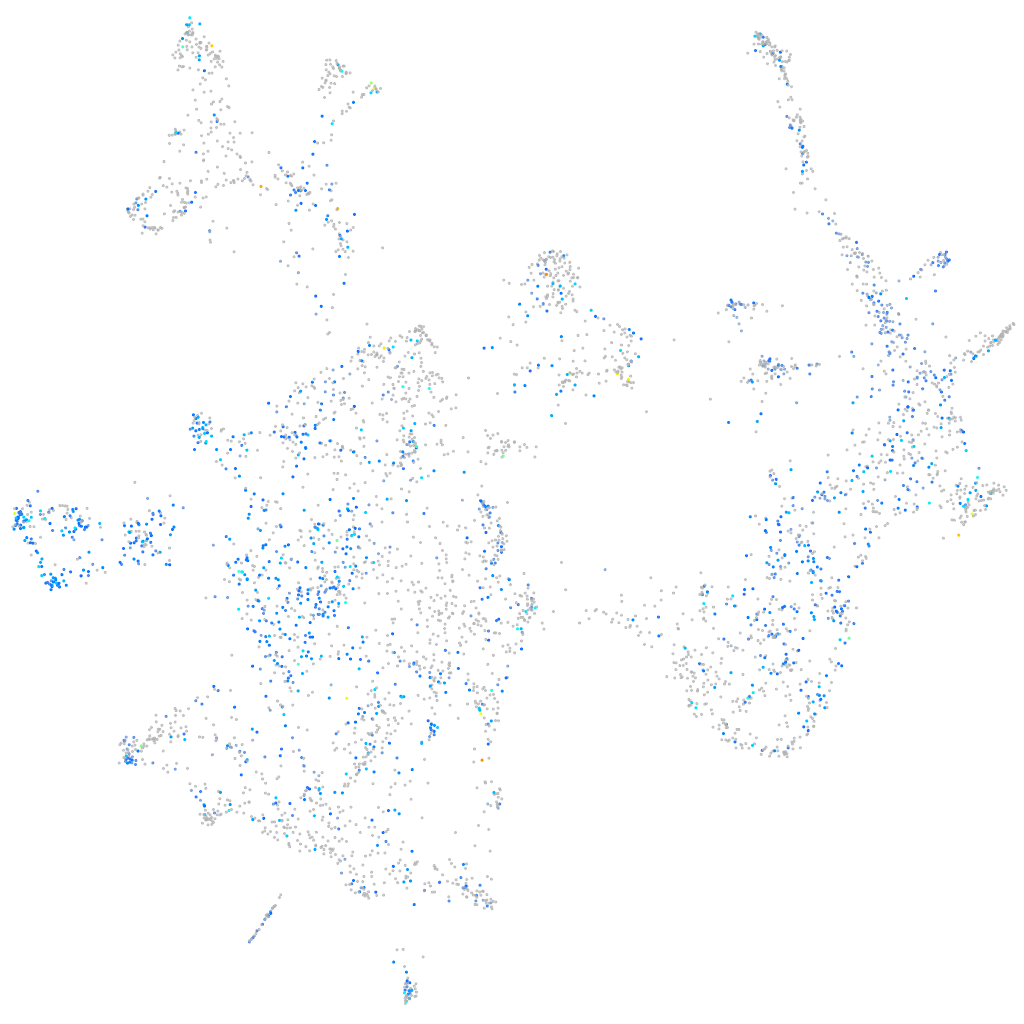
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb2a | 0.278 | COX3 | -0.145 |
| rrm1 | 0.275 | zgc:158463 | -0.135 |
| hmgb2b | 0.275 | mt-co2 | -0.106 |
| hnrnpabb | 0.258 | gapdhs | -0.105 |
| ranbp1 | 0.257 | otofb | -0.098 |
| hmga1a | 0.256 | kif1aa | -0.096 |
| setb | 0.256 | slc17a8 | -0.095 |
| cbx3a | 0.253 | atp1b2b | -0.094 |
| anp32b | 0.252 | gpx2 | -0.094 |
| snrpb | 0.246 | atp2b1a | -0.093 |
| seta | 0.243 | sncb | -0.093 |
| pcna | 0.243 | calml4a | -0.092 |
| hnrnpa0l | 0.242 | s100t | -0.091 |
| alyref | 0.241 | myo7aa | -0.090 |
| snrpd1 | 0.241 | mt-co1 | -0.089 |
| dut | 0.239 | calm1b | -0.089 |
| cnbpa | 0.238 | cabp2b | -0.089 |
| nasp | 0.238 | osbpl1a | -0.088 |
| ddx39ab | 0.237 | ckbb | -0.088 |
| snrpf | 0.237 | apba1a | -0.088 |
| ran | 0.237 | myo6b | -0.087 |
| h3f3a | 0.234 | eno1a | -0.087 |
| khdrbs1a | 0.233 | CR925719.1 | -0.086 |
| nop58 | 0.232 | pvalb1 | -0.086 |
| chaf1a | 0.231 | atp1a3b | -0.086 |
| npm1a | 0.231 | nptna | -0.085 |
| si:ch211-288g17.3 | 0.230 | clic5b | -0.085 |
| h2afvb | 0.230 | pvalb9 | -0.085 |
| hnrnpaba | 0.229 | si:dkey-16p21.8 | -0.085 |
| sumo3b | 0.227 | gb:eh456644 | -0.085 |
| hnrnpa0b | 0.227 | cd164l2 | -0.084 |
| tubb2b | 0.227 | atp6v1aa | -0.083 |
| hmgn2 | 0.225 | atp6v0cb | -0.083 |
| mcm7 | 0.225 | s100s | -0.083 |
| snu13b | 0.225 | anxa5a | -0.083 |